| Eukaryotes | |
|---|---|

| |
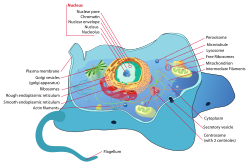
| Eukaryotes and some examples of their diversity – clockwise from top left: Red mason bee, Boletus edulis, Common chimpanzee, Isotricha intestinalis, Persian buttercup, and Volvox carteri | |
| Scientific classification | |
| Domain: | Eukaryota (Chatton, 1925) Whittaker & Margulis, 1978 |
| Supergroups and kingdoms | |
Eukaryotic organisms that cannot be classified under the kingdoms Plantae, Animalia or Fungi are sometimes grouped in the kingdom Protista. | |
Eukaryotes (/juːˈkærioʊt,
Eukaryotes can reproduce both asexually through mitosis and sexually through meiosis and gamete fusion. In mitosis, one cell divides to produce two genetically identical cells. In meiosis, DNA replication is followed by two rounds of cell division to produce four haploid daughter cells. These act as sex cells (gametes). Each gamete has just one set of chromosomes, each a unique mix of the corresponding pair of parental chromosomes resulting from genetic recombination during meiosis.
The domain Eukaryota appears to be monophyletic, and makes up one of the domains of life in the three-domain system. The two other domains, Bacteria and Archaea, are prokaryotes and have none of the above features. Eukaryotes represent a tiny minority of all living things. However, due to their generally much larger size, their collective worldwide biomass is estimated to be about equal to that of prokaryotes. Eukaryotes evolved approximately 1.6–2.1 billion years ago, during the Proterozoic eon.
History
Konstantin Mereschkowski proposed a symbiotic origin for cells with nuclei.
The concept of the eukaryote has been attributed to the French biologist Edouard Chatton (1883-1947). The terms prokaryote and eukaryote were more definitively reintroduced by the Canadian microbiologist Roger Stanier and the Dutch-American microbiologist C. B. van Niel in 1962. In his 1938 work Titres et Travaux Scientifiques, Chatton had proposed the two terms, calling the bacteria prokaryotes and organisms with nuclei in their cells eukaryotes. However he mentioned this in only one paragraph, and the idea was effectively ignored until Chatton's statement was rediscovered by Stanier and van Niel.
In 1905 and 1910, the Russian biologist Konstantin Mereschkowski (1855–1921) argued that plastids were reduced cyanobacteria in a symbiosis with a non-photosynthetic (heterotrophic) host that was itself formed by symbiosis between an amoeba-like host and a bacterium-like cell that formed the nucleus. Plants had thus inherited photosynthesis from cyanobacteria.
In 1967, Lynn Margulis provided microbiological evidence for endosymbiosis as the origin of chloroplasts and mitochondria in eukaryotic cells in her paper, On the origin of mitosing cells. In the 1970s, Carl Woese explored microbial phylogenetics, studying variations in 16S ribosomal RNA. This helped to uncover the origin of the eukaryotes and the symbiogenesis of two important eukaryote organelles, mitochondria and chloroplasts. In 1977, Woese and George Fox introduced a "third form of life", which they called the Archaebacteria; in 1990, Woese, Otto Kandler and Mark L. Wheeler renamed this the Archaea.
In 1979, G. W. Gould and G. J. Dring suggested that the eukaryotic cell's nucleus came from the ability of Gram-positive bacteria to form endospores. In 1987 and later papers, Thomas Cavalier-Smith proposed instead that the membranes of the nucleus and endoplasmic reticulum first formed by infolding a prokaryote's plasma membrane. In the 1990s, several other biologists proposed endosymbiotic origins for the nucleus, effectively reviving Mereschkowsky's theory.
Cell features
Eukaryotic cells are typically much larger than those of prokaryotes having a volume of around 10,000 times greater than the prokaryotic cell. They have a variety of internal membrane-bound structures, called organelles, and a cytoskeleton composed of microtubules, microfilaments, and intermediate filaments, which play an important role in defining the cell's organization and shape. Eukaryotic DNA is divided into several linear bundles called chromosomes, which are separated by a microtubular spindle during nuclear division.Internal membrane
The endomembrane system and its components
Eukaryote cells include a variety of membrane-bound structures, collectively referred to as the endomembrane system. Simple compartments, called vesicles and vacuoles, can form by budding off other membranes. Many cells ingest food and other materials through a process of endocytosis, where the outer membrane invaginates and then pinches off to form a vesicle. It is probable that most other membrane-bound organelles are ultimately derived from such vesicles. Alternatively some products produced by the cell can leave in a vesicle through exocytosis.
The nucleus is surrounded by a double membrane (commonly referred to as a nuclear membrane or nuclear envelope), with pores that allow material to move in and out. Various tube- and sheet-like extensions of the nuclear membrane form the endoplasmic reticulum, which is involved in protein transport and maturation. It includes the rough endoplasmic reticulum where ribosomes are attached to synthesize proteins, which enter the interior space or lumen. Subsequently, they generally enter vesicles, which bud off from the smooth endoplasmic reticulum. In most eukaryotes, these protein-carrying vesicles are released and further modified in stacks of flattened vesicles (cisternae), the Golgi apparatus.
Vesicles may be specialized for various purposes. For instance, lysosomes contain digestive enzymes that break down most biomolecules in the cytoplasm. Peroxisomes are used to break down peroxide, which is otherwise toxic. Many protozoans have contractile vacuoles, which collect and expel excess water, and extrusomes, which expel material used to deflect predators or capture prey. In higher plants, most of a cell's volume is taken up by a central vacuole, which mostly contains water and primarily maintains its osmotic pressure.
Mitochondria and plastids
Simplified structure of a mitochondrion
Mitochondria are organelles found in all but one eukaryote. Mitochondria provide energy to the eukaryote cell by converting sugars into ATP. They have two surrounding membranes, each a phospholipid bi-layer; the inner of which is folded into invaginations called cristae where aerobic respiration takes place.
The outer mitochondrial membrane is freely permeable and allows almost anything to enter into the intermembrane space while the inner mitochondrial membrane is semi permeable so allows only some required things into the mitochondrial matrix.
Mitochondria contain their own DNA, which has close structural similarities to bacterial DNA, and which encodes rRNA and tRNA genes that produce RNA which is closer in structure to bacterial RNA than to eukaryote RNA. They are now generally held to have developed from endosymbiotic prokaryotes, probably proteobacteria.
Some eukaryotes, such as the metamonads such as Giardia and Trichomonas, and the amoebozoan Pelomyxa, appear to lack mitochondria, but all have been found to contain mitochondrion-derived organelles, such as hydrogenosomes and mitosomes, and thus have lost their mitochondria secondarily. They obtain energy by enzymatic action on nutrients absorbed from the environment. The metamonad Monocercomonoides has also acquired, by lateral gene transfer, a cytosolic sulfur mobilisation system which provides the clusters of iron and sulfur required for protein synthesis. The normal mitochondrial iron-sulfur cluster pathway has been lost secondarily.
Plants and various groups of algae also have plastids. Plastids also have their own DNA and are developed from endosymbionts, in this case cyanobacteria. They usually take the form of chloroplasts which, like cyanobacteria, contain chlorophyll and produce organic compounds (such as glucose) through photosynthesis. Others are involved in storing food. Although plastids probably had a single origin, not all plastid-containing groups are closely related. Instead, some eukaryotes have obtained them from others through secondary endosymbiosis or ingestion. The capture and sequestering of photosynthetic cells and chloroplasts occurs in many types of modern eukaryotic organisms and is known as kleptoplasty.
Endosymbiotic origins have also been proposed for the nucleus, and for eukaryotic flagella.
Cytoskeletal structures
Longitudinal section through the flagellum of Chlamydomonas reinhardtii
Many eukaryotes have long slender motile cytoplasmic projections, called flagella, or similar structures called cilia. Flagella and cilia are sometimes referred to as undulipodia, and are variously involved in movement, feeding, and sensation. They are composed mainly of tubulin. These are entirely distinct from prokaryotic flagellae. They are supported by a bundle of microtubules arising from a centriole, characteristically arranged as nine doublets surrounding two singlets. Flagella also may have hairs, or mastigonemes, and scales connecting membranes and internal rods. Their interior is continuous with the cell's cytoplasm.
Microfilamental structures composed of actin and actin binding proteins, e.g., α-actinin, fimbrin, filamin are present in submembraneous cortical layers and bundles, as well. Motor proteins of microtubules, e.g., dynein or kinesin and actin, e.g., myosins provide dynamic character of the network.
Centrioles are often present even in cells and groups that do not have flagella, but conifers and flowering plants have neither. They generally occur in groups that give rise to various microtubular roots. These form a primary component of the cytoskeletal structure, and are often assembled over the course of several cell divisions, with one flagellum retained from the parent and the other derived from it. Centrioles produce the spindle during nuclear division.
The significance of cytoskeletal structures is underlined in the determination of shape of the cells, as well as their being essential components of migratory responses like chemotaxis and chemokinesis. Some protists have various other microtubule-supported organelles. These include the radiolaria and heliozoa, which produce axopodia used in flotation or to capture prey, and the haptophytes, which have a peculiar flagellum-like organelle called the haptonema.
Cell wall
The cells of plants and algae, fungi and most chromalveolates have a cell wall, a layer outside the cell membrane, providing the cell with structural support, protection, and a filtering mechanism. The cell wall also prevents over-expansion when water enters the cell.The major polysaccharides making up the primary cell wall of land plants are cellulose, hemicellulose, and pectin. The cellulose microfibrils are linked via hemicellulosic tethers to form the cellulose-hemicellulose network, which is embedded in the pectin matrix. The most common hemicellulose in the primary cell wall is xyloglucan.
Differences among eukaryotic cells
There are many different types of eukaryotic cells, though animals and plants are the most familiar eukaryotes, and thus provide an excellent starting point for understanding eukaryotic structure. Fungi and many protists have some substantial differences, however.Animal cell
Structure of a typical animal cell
Structure of a typical plant cell
All animals are eukaryotic. Animal cells are distinct from those of other eukaryotes, most notably plants, as they lack cell walls and chloroplasts and have smaller vacuoles. Due to the lack of a cell wall, animal cells can transform into a variety of shapes. A phagocytic cell can even engulf other structures.
Plant cell
Plant cells are quite different from the cells of the other eukaryotic organisms. Their distinctive features are:- A large central vacuole (enclosed by a membrane, the tonoplast), which maintains the cell's turgor and controls movement of molecules between the cytosol and sap
- A primary cell wall containing cellulose, hemicellulose and pectin, deposited by the protoplast on the outside of the cell membrane; this contrasts with the cell walls of fungi, which contain chitin, and the cell envelopes of prokaryotes, in which peptidoglycans are the main structural molecules
- The plasmodesmata, pores in the cell wall that link adjacent cells and allow plant cells to communicate with adjacent cells. Animals have a different but functionally analogous system of gap junctions between adjacent cells.
- Plastids, especially chloroplasts that contain chlorophyll, the pigment that gives plants their green color and allows them to perform photosynthesis
- Bryophytes and seedless vascular plants only have flagellae and centrioles in the sperm cells. Sperm of cycads and Ginkgo are large, complex cells that swim with hundreds to thousands of flagellae.
- Conifers (Pinophyta) and flowering plants (Angiospermae) lack the flagellae and centrioles that are present in animal cells.
Fungal cell
Fungal Hyphae Cells
1- Hyphal wall 2- Septum 3- Mitochondrion 4- Vacuole 5- Ergosterol crystal 6- Ribosome 7- Nucleus 8- Endoplasmic reticulum 9- Lipid body 10- Plasma membrane 11- Spitzenkörper 12- Golgi apparatus
1- Hyphal wall 2- Septum 3- Mitochondrion 4- Vacuole 5- Ergosterol crystal 6- Ribosome 7- Nucleus 8- Endoplasmic reticulum 9- Lipid body 10- Plasma membrane 11- Spitzenkörper 12- Golgi apparatus
The cells of fungi are most similar to animal cells, with the following exceptions:
- A cell wall that contains chitin
- Less definition between cells; the hyphae of higher fungi have porous partitions called septa, which allow the passage of cytoplasm, organelles, and, sometimes, nuclei. Primitive fungi have few or no septa, so each organism is essentially a giant multinucleate supercell; these fungi are described as coenocytic.
- Only the most primitive fungi, chytrids, have flagella.
Other eukaryotic cells
Some groups of eukaryotes have unique organelles, such as the cyanelles (unusual chloroplasts) of the glaucophytes, the haptonema of the haptophytes, or the ejectosomes of the cryptomonads. Other structures, such as pseudopodia, are found in various eukaryote groups in different forms, such as the lobose amoebozoans or the reticulose foraminiferans.Reproduction
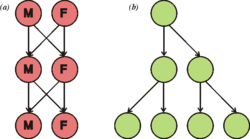
This diagram illustrates the twofold cost of sex. If each individual were to contribute to the same number of offspring (two), (a) the sexual population remains the same size each generation, where the (b) asexual population doubles in size each generation.
Cell division generally takes place asexually by mitosis, a process that allows each daughter nucleus to receive one copy of each chromosome. Most eukaryotes also have a life cycle that involves sexual reproduction, alternating between a haploid phase, where only one copy of each chromosome is present in each cell and a diploid phase, wherein two copies of each chromosome are present in each cell. The diploid phase is formed by fusion of two haploid gametes to form a zygote, which may divide by mitosis or undergo chromosome reduction by meiosis. There is considerable variation in this pattern. Animals have no multicellular haploid phase, but each plant generation can consist of haploid and diploid multicellular phases.
Eukaryotes have a smaller surface area to volume ratio than prokaryotes, and thus have lower metabolic rates and longer generation times.
The evolution of sexual reproduction may be a primordial and fundamental characteristic of eukaryotes. Based on a phylogenetic analysis, Dacks and Roger proposed that facultative sex was present in the common ancestor of all eukaryotes. A core set of genes that function in meiosis is present in both Trichomonas vaginalis and Giardia intestinalis, two organisms previously thought to be asexual. Since these two species are descendants of lineages that diverged early from the eukaryotic evolutionary tree, it was inferred that core meiotic genes, and hence sex, were likely present in a common ancestor of all eukaryotes. Eukaryotic species once thought to be asexual, such as parasitic protozoa of the genus Leishmania, have been shown to have a sexual cycle. Also, evidence now indicates that amoebae, previously regarded as asexual, are anciently sexual and that the majority of present-day asexual groups likely arose recently and independently.
Classification
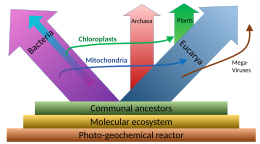
Phylogenetic and symbiogenetic tree of living organisms, showing a view of the origins of eukaryotes & prokaryotes
One hypothesis of eukaryotic relationships. The Opisthokonta group includes both animals (Metazoa) and fungi. Plants (Plantae) are placed in Archaeplastida.
A
pie chart of described eukaryote species (except for Excavata),
together with a tree showing possible relationships between the groups
In antiquity, the two lineages of animals and plants were recognized. They were given the taxonomic rank of Kingdom by Linnaeus. Though he included the fungi with plants with some reservations, it was later realized that they are quite distinct and warrant a separate kingdom, the composition of which was not entirely clear until the 1980s. The various single-cell eukaryotes were originally placed with plants or animals when they became known. In 1830, the German biologist Georg A. Goldfuss coined the word protozoa to refer to organisms such as ciliates, and this group was expanded until it encompassed all single-celled eukaryotes, and given their own kingdom, the Protista, by Ernst Haeckel in 1866. The eukaryotes thus came to be composed of four kingdoms:
The protists were understood to be "primitive forms", and thus an evolutionary grade, united by their primitive unicellular nature. The disentanglement of the deep splits in the tree of life only really started with DNA sequencing, leading to a system of domains rather than kingdoms as top level rank being put forward by Carl Woese, uniting all the eukaryote kingdoms under the eukaryote domain. At the same time, work on the protist tree intensified, and is still actively going on today. Several alternative classifications have been forwarded, though there is no consensus in the field.
Eukaryotes are a clade usually assessed to be sister to Heimdallarchaeota in the Asgard grouping in the Archaea. The basal groupings are the Opimoda, Diphoda, the Discoba, and the Loukozoa. The Eukaryote root is usually assessed to be near or even in Discoba.
A classification produced in 2005 for the International Society of Protistologists, which reflected the consensus of the time, divided the eukaryotes into six supposedly monophyletic 'supergroups'. However, in the same year (2005), doubts were expressed as to whether some of these supergroups were monophyletic, particularly the Chromalveolata, and a review in 2006 noted the lack of evidence for several of the supposed six supergroups. A revised classification in 2012 recognizes five supergroups.
| Archaeplastida (or Primoplantae) | Land plants, green algae, red algae, and glaucophytes |
| SAR supergroup | Stramenopiles (brown algae, diatoms, etc.), Alveolata, and Rhizaria (Foraminifera, Radiolaria, and various other amoeboid protozoa). |
| Excavata | Various flagellate protozoa |
| Amoebozoa | Most lobose amoeboids and slime molds |
| Opisthokonta | Animals, fungi, choanoflagellates, etc. |
There are also smaller groups of eukaryotes whose position is uncertain or seems to fall outside the major groups — in particular, Haptophyta, Cryptophyta, Centrohelida, Telonemia, Picozoa, Apusomonadida, Ancyromonadida, Breviatea, and the genus Collodictyon. Overall, it seems that, although progress has been made, there are still very significant uncertainties in the evolutionary history and classification of eukaryotes. As Roger & Simpson said in 2009 "with the current pace of change in our understanding of the eukaryote tree of life, we should proceed with caution."
In an article published in Nature Microbiology in April 2016 the authors, "reinforced once again that the life we see around us – plants, animals, humans and other so-called eukaryotes – represent a tiny percentage of the world's biodiversity." They classified eukaryote "based on the inheritance of their information systems as opposed to lipid or other cellular structures." Jillian F. Banfield of the University of California, Berkeley and fellow scientists used a super computer to generate a diagram of a new tree of life based on DNA from 3000 species including 2,072 known species and 1,011 newly reported microbial organisms, whose DNA they had gathered from diverse environments. As the capacity to sequence DNA became easier, Banfield and team were able to do metagenomic sequencing—"sequencing whole communities of organisms at once and picking out the individual groups based on their genes alone."
Phylogeny
The rRNA trees constructed during the 1980s and 1990s left most eukaryotes in an unresolved "crown" group (not technically a true crown), which was usually divided by the form of the mitochondrial cristae; see crown eukaryotes. The few groups that lack mitochondria branched separately, and so the absence was believed to be primitive; but this is now considered an artifact of long-branch attraction, and they are known to have lost them secondarily.As of 2011, there is widespread agreement that the Rhizaria belong with the Stramenopiles and the Alveolata, in a clade dubbed the SAR supergroup, so that Rhizaria is not one of the main eukaryote groups; also that the Amoebozoa and Opisthokonta are each monophyletic and form a clade, often called the unikonts. Beyond this, there does not appear to be a consensus.
It has been estimated that there may be 75 distinct lineages of eukaryotes. Most of these lineages are protists.
The known eukaryote genome sizes vary from 8.2 megabases (Mb) in Babesia bovis to 112,000–220,050 Mb in the dinoflagellate Prorocentrum micans, showing that the genome of the ancestral eukaryote has undergone considerable variation during its evolution. The last common ancestor of all eukaryotes is believed to have been a phagotrophic protist with a nucleus, at least one centriole and cilium, facultatively aerobic mitochondria, sex (meiosis and syngamy), a dormant cyst with a cell wall of chitin and/or cellulose and peroxisomes. Later endosymbiosis led to the spread of plastids in some lineages.
Five supergroups
A global tree of eukaryotes from a consensus of phylogenetic evidence (in particular, phylogenomics), rare genomic signatures, and morphological characteristics is presented in Adl et al. 2012 and Burki 2014/2016 with the Cryptophyta and picozoa having emerged within the Archaeplastida. A similar inclusion of Glaucophyta, Cryptista (and also, unusually, Haptista) has also been made.In some analyses, the Hacrobia group (Haptophyta + Cryptophyta) is placed next to Archaeplastida, but in other ones it is nested inside the Archaeplastida. However, several recent studies have concluded that Haptophyta and Cryptophyta do not form a monophyletic group. The former could be a sister group to the SAR group, the latter cluster with the Archaeplastida (plants in the broad sense).\
The division of the eukaryotes into two primary clades, bikonts (Archaeplastida + SAR + Excavata) and unikonts (Amoebozoa + Opisthokonta), derived from an ancestral biflagellar organism and an ancestral uniflagellar organism, respectively, had been suggested earlier. A 2012 study produced a somewhat similar division, although noting that the terms "unikonts" and "bikonts" were not used in the original sense.
A highly converged and congruent set of trees appears in Derelle et al (2015), Ren et al (2016), Yang et al (2017) and Cavalier-Smith (2015) including the supplementary information, resulting in a more conservative and consolidated tree. It is combined with some results from Cavalier-Smith for the basal Opimoda. The main remaining controversies are the root, and the exact positioning of the Rhodophyta and the bikonts Rhizaria, Haptista, Cryptista, Picozoa and Telonemia, many of which may be endosymbyotic eukaryote-eukaryote hybrids. Archeaplastida developed the Chloroplasts probably by endosymbiosis of an ancestor related to a currently extant cyanobacterium, Gloeomargarita lithophora.
Cavalier-Smith's tree
Thomas Cavalier-Smith 2010, 2013, 2014, 2017, and 2018 places the eukaryotic tree's root between Excavata (with ventral feeding groove supported by a microtubular root) and the grooveless Euglenozoa, and monophyletic Chromista, correlated to a single endosymbyotic event of capturing a red-algae. He et al specifically supports rooting eukaryotic tree between a monophyletic Discoba (Discicristata + Jakobida) and a Amorphea-Diaphoretickes clade.Origin of eukaryotes
The three-domains tree and the Eocyte hypothesis
Phylogenetic tree showing a possible relationship between the eukaryotes and other forms of life; eukaryotes are colored red, archaea green and bacteria blue.
Eocyte tree.
Fossils
The origin of the eukaryotic cell is a milestone in the evolution of life, since eukaryotes include all complex cells and almost all multicellular organisms. The timing of this series of events is hard to determine; Knoll (2006) suggests they developed approximately 1.6–2.1 billion years ago. Some acritarchs are known from at least 1.65 billion years ago, and the possible alga Grypania has been found as far back as 2.1 billion years ago. The Geosiphon-like fossil fungus Diskagma has been found in paleosols 2.2 billion years old.Organized living structures have been found in the black shales of the Palaeoproterozoic Francevillian B Formation in Gabon, dated at 2.1 billion years old. Eukaryotic life could have evolved at that time. Fossils that are clearly related to modern groups start appearing an estimated 1.2 billion years ago, in the form of a red alga, though recent work suggests the existence of fossilized filamentous algae in the Vindhya basin dating back perhaps to 1.6 to 1.7 billion years ago.
Biomarkers suggest that at least stem eukaryotes arose even earlier. The presence of steranes in Australian shales indicates that eukaryotes were present in these rocks dated at 2.7 billion years old, although it was suggested they could originate from samples contamination.
Whenever their origins, eukaryotes may not have become ecologically dominant until much later; a massive uptick in the zinc composition of marine sediments 800 million years ago has been attributed to the rise of substantial populations of eukaryotes, which preferentially consume and incorporate zinc relative to prokaryotes.
Relationship to Archaea
The nuclear DNA and genetic machinery of eukaryotes is more similar to Archaea than Bacteria, leading to a controversial suggestion that eukaryotes should be grouped with Archaea in the clade Neomura. In other respects, such as membrane composition, eukaryotes are similar to Bacteria. Three main explanations for this have been proposed:- Eukaryotes resulted from the complete fusion of two or more cells, wherein the cytoplasm formed from a eubacterium, and the nucleus from an archaeon, from a virus, or from a pre-cell.
- Eukaryotes developed from Archaea, and acquired their eubacterial characteristics through the endosymbiosis of a proto-mitochondrion of eubacterial origin.
- Eukaryotes and Archaea developed separately from a modified eubacterium.
Diagram
of the origin of life with the Eukaryotes appearing early, not derived
from Prokaryotes, as proposed by Richard Egel in 2012. This view implies
that the UCA was relatively large and complex.
- The chronocyte hypothesis postulates that a primitive eukaryotic cell was formed by the endosymbiosis of both archaea and bacteria by a third type of cell, termed a chronocyte.
- The universal common ancestor (UCA) of the current tree of life was a complex organism that survived a mass extinction event rather than an early stage in the evolution of life. Eukaryotes and in particular akaryotes (Bacteria and Archaea) evolved through reductive loss, so that similarities result from differential retention of original features.
In recent years, most researchers have favoured either the three domains (3D) or the eocyte hypotheses. An rRNA analyses supports the eocyte scenario, apparently with the Eukaryote root in Excavata.
In this scenario, the Asgard group is seen as a sister taxon of the TACK group, which comprises Crenarchaeota (formerly named eocytes), Thaumarchaeota, and others.
In 2017, there has been significant pushback against this scenario, arguing that the eukaryotes did not emerge within the Archaea. Cunha et al. produced analyses supporting the three domains (3D) or Woese hypothesis (2 in the table above) and rejecting the eocyte hypothesis (4 above). Harish and Kurland found strong support for the earlier two empires (2D) or Mayr hypothesis (1 in the table above), based on analyses of the coding sequences of protein domains. They rejected the eocyte hypothesis as the least likely. A possible interpretation of their analysis is that the universal common ancestor (UCA) of the current tree of life was a complex organism that survived an evolutionary bottleneck, rather than a simpler organism arising early in the history of life.
Endomembrane system and mitochondria
The origins of the endomembrane system and mitochondria are also unclear. The phagotrophic hypothesis proposes that eukaryotic-type membranes lacking a cell wall originated first, with the development of endocytosis, whereas mitochondria were acquired by ingestion as endosymbionts. The syntrophic hypothesis proposes that the proto-eukaryote relied on the proto-mitochondrion for food, and so ultimately grew to surround it. Here the membranes originated after the engulfment of the mitochondrion, in part thanks to mitochondrial genes (the hydrogen hypothesis is one particular version).In a study using genomes to construct supertrees, Pisani et al. (2007) suggest that, along with evidence that there was never a mitochondrion-less eukaryote, eukaryotes evolved from a syntrophy between an archaea closely related to Thermoplasmatales and an α-proteobacterium, likely a symbiosis driven by sulfur or hydrogen. The mitochondrion and its genome is a remnant of the α-proteobacterial endosymbiont.
Hypotheses
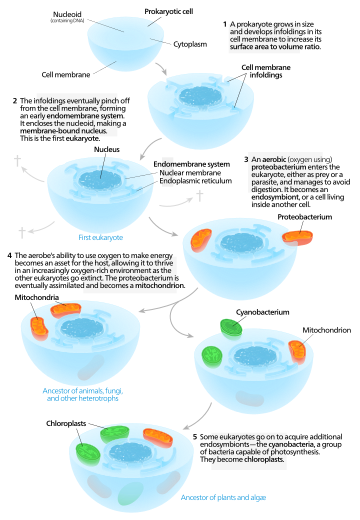
Different hypotheses have been proposed as to how eukaryotic cells came into existence. These hypotheses can be classified into two distinct classes – autogenous models and chimeric models.Autogenous models
Autogenous models propose that a proto-eukaryotic cell containing a nucleus existed first, and later acquired mitochondria. According to this model, a large prokaryote developed invaginations in its plasma membrane in order to obtain enough surface area to service its cytoplasmic volume. As the invaginations differentiated in function, some became separate compartments—giving rise to the endomembrane system, including the endoplasmic reticulum, golgi apparatus, nuclear membrane, and single membrane structures such as lysosomes. Mitochondria are proposed to come from the endosymbiosis of an aerobic proteobacterium, and it is assumed that all the eukaryotic lineages that did not acquire mitochondria became extinct. Chloroplasts came about from another endosymbiotic event involving cyanobacteria. Since all eukaryotes have mitochondria, but not all have chloroplasts, the serial endosymbiosis theory proposes that mitochondria came first.Chimeric models
Chimeric models claim that two prokaryotic cells existed initially – an archaeon and a bacterium. These cells underwent a merging process, either by a physical fusion or by endosymbiosis, thereby leading to the formation of a eukaryotic cell. Within these chimeric models, some studies further claim that mitochondria originated from a bacterial ancestor while others emphasize the role of endosymbiotic processes behind the origin of mitochondria.Based on the process of mutualistic symbiosis, the hypotheses can be categorized as – the serial endosymbiotic theory (SET), the hydrogen hypothesis (mostly a process of symbiosis where hydrogen transfer takes place among different species), and the syntrophy hypothesis.
According to serial endosymbiotic theory (championed by Lynn Margulis), a union between a motile anaerobic bacterium (like Spirochaeta) and a thermoacidophilic crenarchaeon (like Thermoplasma which is sulfidogenic in nature) gave rise to the present day eukaryotes. This union established a motile organism capable of living in the already existing acidic and sulfurous waters. Oxygen is known to cause toxicity to organisms that lack the required metabolic machinery. Thus, the archaeon provided the bacterium with a highly beneficial reduced environment (sulfur and sulfate were reduced to sulfide). In microaerophilic conditions, oxygen was reduced to water thereby creating a mutual benefit platform. The bacterium on the other hand, contributed the necessary fermentation products and electron acceptors along with its motility feature to the archaeon thereby gaining a swimming motility for the organism. From a consortium of bacterial and archaeal DNA originated the nuclear genome of eukaryotic cells. Spirochetes gave rise to the motile features of eukaryotic cells. Endosymbiotic unifications of the ancestors of alpha-proteobacteria and cyanobacteria, led to the origin of mitochondria and plastids respectively. For example, Thiodendron has been known to have originated via an ectosymbiotic process based on a similar syntrophy of sulfur existing between the two types of bacteria – Desulphobacter and Spirochaeta. However, such an association based on motile symbiosis have never been observed practically. Also there is no evidence of archaeans and spirochetes adapting to intense acid-based environments.
In the hydrogen hypothesis, the symbiotic linkage of an anaerobic and autotrophic methanogenic archaeon (host) with an alpha-proteobacterium (the symbiont) gave rise to the eukaryotes. The host utilized hydrogen (H2) and carbon dioxide (CO2) to produce methane while the symbiont, capable of aerobic respiration, expelled H2 and CO2 as byproducts of anaerobic fermentation process. The host's methanogenic environment worked as a sink for H2, which resulted in heightened bacterial fermentation. Endosymbiotic gene transfer (EGT) acted as a catalyst for the host to acquire the symbionts' carbohydrate metabolism and turn heterotrophic in nature. Subsequently, the host's methane forming capability was lost. Thus, the origins of the heterotrophic organelle (symbiont) are identical to the origins of the eukaryotic lineage. In this hypothesis, the presence of H2 represents the selective force that forged eukaryotes out of prokaryotes.
The syntrophy hypothesis was developed in contrast to the hydrogen hypothesis and proposes the existence of two symbiotic events. According to this theory, the origin of eukaryotic cells was based on metabolic symbiosis (syntrophy) between a methanogenic archaeon and a delta-proteobacterium. This syntrophic symbiosis was initially facilitated by H2 transfer between different species under anaerobic environments. In earlier stages, an alpha-proteobacterium became a member of this integration, and later developed into the mitochondrion. Gene transfer from a delta-proteobacterium to an archaeon led to the methanogenic archaeon developing into a nucleus. The archaeon constituted the genetic apparatus, while the delta-proteobacterium contributed towards the cytoplasmic features. This theory incorporates two selective forces at the time of nucleus evolution – (a) presence of metabolic partitioning to avoid the harmful effects of the co-existence of anabolic and catabolic cellular pathways, and (b) prevention of abnormal protein biosynthesis due to a vast spread of introns in the archaeal genes after acquiring the mitochondrion and losing methanogenesis.