Gene delivery is the process of introducing foreign genetic material, such as DNA or RNA, into host cells. Genetic material must reach the nucleus of the host cell to induce gene expression. Successful gene delivery requires the foreign genetic material to
remain stable within the host cell and can either integrate into the genome or replicate independently of it. This requires foreign DNA to be synthesized as part of a vector, which is designed to enter the desired host cell and deliver the transgene to that cell's genome.
Vectors utilized as the method for gene delivery can be divided into
two categories, recombinant viruses and synthetic vectors (viral and
non-viral).
In complex multicellular eukaryotes (more specifically Weissmanists), if the transgene is incorporated into the host's germline cells, the resulting host cell can pass the transgene to its progeny. If the transgene is incorporated into somatic cells, the transgene will stay with the somatic cell line, and thus its host organism.[6]
Gene delivery is a necessary step in gene therapy for the introduction or silencing of a gene to promote a therapeutic outcome in patients and also has applications in the genetic modification of crops. There are many different methods of gene delivery for various types of cells and tissues. [7]
In complex multicellular eukaryotes (more specifically Weissmanists), if the transgene is incorporated into the host's germline cells, the resulting host cell can pass the transgene to its progeny. If the transgene is incorporated into somatic cells, the transgene will stay with the somatic cell line, and thus its host organism.[6]
Gene delivery is a necessary step in gene therapy for the introduction or silencing of a gene to promote a therapeutic outcome in patients and also has applications in the genetic modification of crops. There are many different methods of gene delivery for various types of cells and tissues. [7]
History
Viral based vectors emerged in the 1980's as a tool for transgene expression. In 1983, Siegel described the use of viral vectors in plant transgene expression although viral manipulation via cDNA cloning was not yet available. [8] The first virus to be used as a vaccine vector was the vaccinia virus in 1984 as a way to protect chimpanzees against Hepatitis B. [9] Non-viral gene delivery was first reported on in 1943 by Avery et al. who showed cellular phenotype change via Exogenous DNA exposure. [10]Methods
Electroporator
with square wave and exponential decay waveforms for in vitro, in vivo,
adherent cell and 96 well electroporation applications. Manufactured by
BTX Harvard Apparatus, Holliston MA USA.
Non-viral Delivery
Non-viral based gene delivery encompasses chemical and physical delivery methods.[11] Non-viral methods of gene delivery are less likely to induce an immune response, compared to viral vectors. They are more cost-efficient and can deliver larger sizes of genetic material. A draw back of non-viral gene delivery is low efficiency.[11]Chemical
Non-viral chemical based methods of gene delivery uses natural or synthetic compounds to form particles that facilitate the transfer of genes into cells.[12] These synthetic vectors have the ability to electrostatically bind DNA or RNA and compact the genetic information to accommodate larger genetic transfers.[13] Non-viral chemical vectors enter cells by endocytosis and can protect genetic material from degradation.[11]Two common non-viral vectors are liposomes and polymers. Liposome-based non-viral vectors uses liposomes to facilitate gene delivery by the formation of lipoplexes. Lipoplexes are spontaneously formed when positively charged liposomes complex with negatively charged DNA.[12] Polymer-based non-viral vectors uses polymers to interact with DNA and form polyplexes.[11]
The use of engineered organic nanoparticles is another non-viral approach for gene delivery.[14]
Physical
Artificial non-viral gene delivery can be mediated by physical methods which uses force to introduce genetic material through the cell membrane.[12]Physical methods of gene delivery include:[12]
- Needle injection - Direct injection of genetic material using a needle
- Ballistic DNA injection - Gold coated DNA particles are forced into cells
- Electroporation -Electric pulses create pores in a cell membrane to allow entry of genetic material
- Sonoporation - Sound waves create pores in a cell membrane to allow entry of genetic material
- Photoporation - Laser pulses create pores in a cell membrane to allow entry of genetic material
- Magnetofection - Magnetic particles complexed with DNA and an external magnetic field concentrate nucleic acid particles into target cells
- Hydroporation - Hydrodynamic capillary effect manipulates cell permeability
Viral Delivery
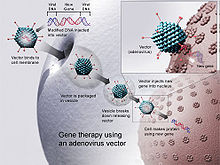
Foreign DNA being transduced into the host cell through an adenovirus vector.
Virus mediated gene delivery utilizes the ability of a virus to inject its DNA inside a host cell and takes advantage of the virus' own ability to replicate and implement their own genetic material. Transduction is the process through which DNA is injected into the host cell and inserted into its genome [needs citation]. Viruses are a particularly effective form of gene delivery because the structure of the virus prevents degradation via lysosomes of the DNA it is delivering to the nucleus of the host cell.[15] In gene therapy a gene that is intended for delivery is packaged into a replication-deficient viral particle to form a viral vector.[16] Viruses used for gene therapy to date include retrovirus, adenovirus, adeno-associated virus and herpes simplex virus. However, there are drawbacks to using viruses to deliver genes into cells. Viruses can only deliver very small pieces of DNA into the cells, it is labor-intensive and there are risks of random insertion sites, cytophathic effects and mutagenesis.[17]
Viral vector based gene delivery uses a viral vector to deliver genetic material to the host cell. This is done by using a virus that contains the desired gene and removing the part of the viruses genome that is infectious.[18] Viruses are efficient at delivering genetic material to the host cell's nucleus, which is vital for replication.[15]
RNA-based viral vectors
RNA-based viruses were developed because of the ability to transcribe directly from infectious RNA transcripts. RNA vectors are quickly expressed and expressed in the targeted form since no processing is required. Gene integration leads to long-term transgene expression but RNA-based delivery is usually transient and not permanent.[2] Some retroviral vectors include: Oncoretroviral vectors, Lentiviral vector in gene therapy, Human foamy virus[2]DNA-based viral vectors
DNA-based viral vectors are usually longer lasting with the possibility of integrating into the genome. Some DNA-based viral vectors include: Adenoviridae, Adeno-associated virus, Herpes simplex virus[2]Applications
Gene Therapy
Several of the methods used to facilitate gene delivery have applications for therapeutic purposes. Gene therapy utilizes gene delivery to deliver genetic material with the goal of treating a disease or condition in the cell. Gene delivery in therapeutic settings utilizes non-immunogenic vectors capable of cell specificity that can deliver an adequate amount of transgene expression to cause the desired effect.[19]Advances in genomics have enabled a variety of new methods and gene targets to be identified for possible applications. DNA microarrays used in a variety of next-gen sequencing can identify thousands of genes simultaneously, with analytical software looking at gene expression patterns, and orthologous genes in model species to identify function.[20] This has allowed a variety of possible vectors to be identified for use in gene therapy. As a method for creating a new class of vaccine, gene delivery has been utilized to generate a hybrid biosynthetic vector to deliver a possible vaccine. This vector overcomes traditional barriers to gene delivery by combining E. coli with a synthetic polymer to create a vector that maintains plasmid DNA while having an increased ability to avoid degradation by target cell lysosomes.