Molecular evolution is the process of change in the sequence composition of cellular molecules such as DNA, RNA, and proteins across generations. The field of molecular evolution uses principles of evolutionary biology and population genetics to explain patterns in these changes. Major topics in molecular evolution concern the rates and impacts of single nucleotide changes, neutral evolution vs. natural selection, origins of new genes, the genetic nature of complex traits, the genetic basis of speciation, evolution of development, and ways that evolutionary forces influence genomic and phenotypic changes.
History
The history of molecular evolution starts in the early 20th century with comparative biochemistry, and the use of "fingerprinting" methods such as immune assays, gel electrophoresis and paper chromatography in the 1950s to explore homologous proteins.[1][2] The field of molecular evolution came into its own in the 1960s and 1970s, following the rise of molecular biology. The advent of protein sequencing allowed molecular biologists to create phylogenies based on sequence comparison, and to use the differences between homologous sequences as a molecular clock to estimate the time since the last universal common ancestor.[1] In the late 1960s, the neutral theory of molecular evolution provided a theoretical basis for the molecular clock,[3] though both the clock and the neutral theory were controversial, since most evolutionary biologists held strongly to panselectionism, with natural selection as the only important cause of evolutionary change. After the 1970s, nucleic acid sequencing allowed molecular evolution to reach beyond proteins to highly conserved ribosomal RNA sequences, the foundation of a reconceptualization of the early history of life.[1]Forces in molecular evolution
The content and structure of a genome is the product of the molecular and population genetic forces which act upon that genome. Novel genetic variants will arise through mutation and will spread and be maintained in populations due to genetic drift or natural selection.Mutation

This hedgehog has no pigmentation due to a mutation.
Mutations are permanent, transmissible changes to the genetic material (DNA or RNA) of a cell or virus. Mutations result from errors in DNA replication during cell division and by exposure to radiation, chemicals, and other environmental stressors, or viruses and transposable elements. Most mutations that occur are single nucleotide polymorphisms which modify single bases of the DNA sequence, resulting in point mutations. Other types of mutations modify larger segments of DNA and can cause duplications, insertions, deletions, inversions, and translocations.
Most organisms display a strong bias in the types of mutations that occur with strong influence in GC-content. Transitions (A ↔ G or C ↔ T) are more common than transversions (purine (adenine or guanine)) ↔ pyrimidine (cytosine or thymine, or in RNA, uracil))[4] and are less likely to alter amino acid sequences of proteins.
Mutations are stochastic and typically occur randomly across genes. Mutation rates for single nucleotide sites for most organisms are very low, roughly 10−9 to 10−8 per site per generation, though some viruses have higher mutation rates on the order of 10−6 per site per generation. Among these mutations, some will be neutral or beneficial and will remain in the genome unless lost via genetic drift, and others will be detrimental and will be eliminated from the genome by natural selection.
Because mutations are extremely rare, they accumulate very slowly across generations. While the number of mutations which appears in any single generation may vary, over very long time periods they will appear to accumulate at a regular pace. Using the mutation rate per generation and the number of nucleotide differences between two sequences, divergence times can be estimated effectively via the molecular clock.
Recombination
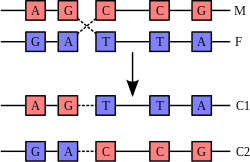
Recombination involves the breakage and rejoining of two chromosomes (M
and F) to produce two re-arranged chromosomes (C1 and C2).
Recombination is a process that results in genetic exchange between chromosomes or chromosomal regions. Recombination counteracts physical linkage between adjacent genes, thereby reducing genetic hitchhiking. The resulting independent inheritance of genes results in more efficient selection, meaning that regions with higher recombination will harbor fewer detrimental mutations, more selectively favored variants, and fewer errors in replication and repair. Recombination can also generate particular types of mutations if chromosomes are misaligned.
Gene conversion
Gene conversion is a type of recombination that is the product of DNA repair where nucleotide damage is corrected using an homologous genomic region as a template. Damaged bases are first excised, the damaged strand is then aligned with an undamaged homolog, and DNA synthesis repairs the excised region using the undamaged strand as a guide. Gene conversion is often responsible for homogenizing sequences of duplicate genes over long time periods, reducing nucleotide divergence.Genetic drift
Genetic drift is the change of allele frequencies from one generation to the next due to stochastic effects of random sampling in finite populations. Some existing variants have no effect on fitness and may increase or decrease in frequency simply due to chance. "Nearly neutral" variants whose selection coefficient is close to a threshold value of 1 / the effective population size will also be affected by chance as well as by selection and mutation. Many genomic features have been ascribed to accumulation of nearly neutral detrimental mutations as a result of small effective population sizes.[5] With a smaller effective population size, a larger variety of mutations will behave as if they are neutral due to inefficiency of selection.Selection
Selection occurs when organisms with greater fitness, i.e. greater ability to survive or reproduce, are favored in subsequent generations, thereby increasing the instance of underlying genetic variants in a population. Selection can be the product of natural selection, artificial selection, or sexual selection. Natural selection is any selective process that occurs due to the fitness of an organism to its environment. In contrast sexual selection is a product of mate choice and can favor the spread of genetic variants which act counter to natural selection but increase desirability to the opposite sex or increase mating success. Artificial selection, also known as selective breeding, is imposed by an outside entity, typically humans, in order to increase the frequency of desired traits.The principles of population genetics apply similarly to all types of selection, though in fact each may produce distinct effects due to clustering of genes with different functions in different parts of the genome, or due to different properties of genes in particular functional classes. For instance, sexual selection could be more likely to affect molecular evolution of the sex chromosomes due to clustering of sex specific genes on the X, Y, Z or W.
Selection can operate at the gene level at the expense of organismal fitness, resulting in a selective advantage for selfish genetic elements in spite of a host cost. Examples of such selfish elements include transposable elements, meiotic drivers, killer X chromosomes, selfish mitochondria, and self-propagating introns. (See Intragenomic conflict.)
Genome architecture
Genome size
Genome size is influenced by the amount of repetitive DNA as well as number of genes in an organism. The C-value paradox refers to the lack of correlation between organism 'complexity' and genome size. Explanations for the so-called paradox are two-fold. First, repetitive genetic elements can comprise large portions of the genome for many organisms, thereby inflating DNA content of the haploid genome. Secondly, the number of genes is not necessarily indicative of the number of developmental stages or tissue types in an organism. An organism with few developmental stages or tissue types may have large numbers of genes that influence non-developmental phenotypes, inflating gene content relative to developmental gene families.Neutral explanations for genome size suggest that when population sizes are small, many mutations become nearly neutral. Hence, in small populations repetitive content and other 'junk' DNA can accumulate without placing the organism at a competitive disadvantage. There is little evidence to suggest that genome size is under strong widespread selection in multicellular eukaryotes. Genome size, independent of gene content, correlates poorly with most physiological traits and many eukaryotes, including mammals, harbor very large amounts of repetitive DNA.
However, birds likely have experienced strong selection for reduced genome size, in response to changing energetic needs for flight. Birds, unlike humans, produce nucleated red blood cells, and larger nuclei lead to lower levels of oxygen transport. Bird metabolism is far higher than that of mammals, due largely to flight, and oxygen needs are high. Hence, most birds have small, compact genomes with few repetitive elements. Indirect evidence suggests that non-avian theropod dinosaur ancestors of modern birds [6] also had reduced genome sizes, consistent with endothermy and high energetic needs for running speed. Many bacteria have also experienced selection for small genome size, as time of replication and energy consumption are so tightly correlated with fitness.
Repetitive elements
Transposable elements are self-replicating, selfish genetic elements which are capable of proliferating within host genomes. Many transposable elements are related to viruses, and share several proteins in common.Chromosome number and organization
The number of chromosomes in an organism's genome also does not necessarily correlate with the amount of DNA in its genome. The ant Myrmecia pilosula has only a single pair of chromosomes[7] whereas the Adders-tongue fern Ophioglossum reticulatum has up to 1260 chromosomes.[8] Cilliate genomes house each gene in individual chromosomes, resulting in a genome which is not physically linked. Reduced linkage through creation of additional chromosomes should effectively increase the efficiency of selection.Changes in chromosome number can play a key role in speciation, as differing chromosome numbers can serve as a barrier to reproduction in hybrids. Human chromosome 2 was created from a fusion of two chimpanzee chromosomes and still contains central telomeres as well as a vestigial second centromere. Polyploidy, especially allopolyploidy, which occurs often in plants, can also result in reproductive incompatibilities with parental species. Agrodiatus blue butterflies have diverse chromosome numbers ranging from n=10 to n=134 and additionally have one of the highest rates of speciation identified to date.[9]
Gene content and distribution
Different organisms house different numbers of genes within their genomes as well as different patterns in the distribution of genes throughout the genome. Some organisms, such as most bacteria, Drosophila, and Arabidopsis have particularly compact genomes with little repetitive content or non-coding DNA. Other organisms, like mammals or maize, have large amounts of repetitive DNA, long introns, and substantial spacing between different genes. The content and distribution of genes within the genome can influence the rate at which certain types of mutations occur and can influence the subsequent evolution of different species. Genes with longer introns are more likely to recombine due to increased physical distance over the coding sequence. As such, long introns may facilitate ectopic recombination, and result in higher rates of new gene formation.Organelles
In addition to the nuclear genome, endosymbiont organelles contain their own genetic material typically as circular plasmids. Mitochondrial and chloroplast DNA varies across taxa, but membrane-bound proteins, especially electron transport chain constituents are most often encoded in the organelle. Chloroplasts and mitochondria are maternally inherited in most species, as the organelles must pass through the egg. In a rare departure, some species of mussels are known to inherit mitochondria from father to son.Origins of new genes
New genes arise from several different genetic mechanisms including gene duplication, de novo origination, retrotransposition, chimeric gene formation, recruitment of non-coding sequence, and gene truncation.Gene duplication initially leads to redundancy. However, duplicated gene sequences can mutate to develop new functions or specialize so that the new gene performs a subset of the original ancestral functions. In addition to duplicating whole genes, sometimes only a domain or part of a protein is duplicated so that the resulting gene is an elongated version of the parental gene.
Retrotransposition creates new genes by copying mRNA to DNA and inserting it into the genome. Retrogenes often insert into new genomic locations, and often develop new expression patterns and functions.
Chimeric genes form when duplication, deletion, or incomplete retrotransposition combine portions of two different coding sequences to produce a novel gene sequence. Chimeras often cause regulatory changes and can shuffle protein domains to produce novel adaptive functions.
De novo origin. Novel genes can also arise from previously non-coding DNA.[10] For instance, Levine and colleagues reported the origin of five new genes in the D. melanogaster genome from noncoding DNA.[11][12] Similar de novo origin of genes has been also shown in other organisms such as yeast,[13] rice[14] and humans.[15] De novo genes may evolve from transcripts that are already expressed at low levels.[16] Mutation of a stop codon to a regular codon or a frameshift may cause an extended protein that includes a previously non-coding sequence.
De novo evolution of genes can also be simulated in the laboratory. Donnelly et al. have shown that semi-random gene sequences can be selected for specific functions. More specifically, they selected sequences from a library that could complement a gene deletion in E. coli. The deleted gene encodes ferric enterobactin esterase (Fes), which releases iron from an iron chelator, enterobactin. While Fes is a 400 amino acid protein, the newly selected gene was only 100 amino acids in length and unrelated in sequence to Fes.[17]
In vitro molecular evolution experiments
Principles of molecular evolution have also been discovered, and others elucidated and tested using experimentation involving amplification, variation and selection of rapidly proliferating and genetically varying molecular species outside cells. Since the pioneering work of Sol Spiegelmann in 1967 [ref], involving RNA that replicates itself with the aid of an enzyme extracted from the Qß virus [ref], several groups (such as Kramers [ref] and Biebricher/Luce/Eigen [ref]) studied mini and micro variants of this RNA in the 1970s and 1980s that replicate on the timescale of seconds to a minute, allowing hundreds of generations with large population sizes (e.g. 10^14 sequences) to be followed in a single day of experimentation. The chemical kinetic elucidation of the detailed mechanism of replication [ref, ref] meant that this type of system was the first molecular evolution system that could be fully characterised on the basis of physical chemical kinetics, later allowing the first models of the genotype to phenotype map based on sequence dependent RNA folding and refolding to be produced [ref, ref]. Subject to maintaining the function of the multicomponent Qß enzyme, chemical conditions could be varied significantly, in order to study the influence of changing environments and selection pressures [ref]. Experiments with in vitro RNA quasi species included the characterisation of the error threshold for information in molecular evolution [ref], the discovery of de novo evolution [ref] leading to diverse replicating RNA species and the discovery of spatial travelling waves as ideal molecular evolution reactors [ref, ref]. Later experiments employed novel combinations of enzymes to elucidate novel aspects of interacting molecular evolution involving population dependent fitness, including work with artificially designed molecular predator prey and cooperative systems of multiple RNA and DNA [ref, ref]. Special evolution reactors were designed for these studies, starting with serial transfer machines, flow reactors such as cell-stat machines, capillary reactors, and microreactors including line flow reactors and gel slice reactors. These studies were accompanied by theoretical developments and simulations involving RNA folding and replication kinetics that elucidated the importance of the correlation structure between distance in sequence space and fitness changes [ref], including the role of neutral networks and structural ensembles in evolutionary optimisation.Molecular phylogenetics
Molecular systematics is the product of the traditional fields of systematics and molecular genetics. It uses DNA, RNA, or protein sequences to resolve questions in systematics, i.e. about their correct scientific classification or taxonomy from the point of view of evolutionary biology.Molecular systematics has been made possible by the availability of techniques for DNA sequencing, which allow the determination of the exact sequence of nucleotides or bases in either DNA or RNA. At present it is still a long and expensive process to sequence the entire genome of an organism, and this has been done for only a few species. However, it is quite feasible to determine the sequence of a defined area of a particular chromosome. Typical molecular systematic analyses require the sequencing of around 1000 base pairs.
The driving forces of evolution
Depending on the relative importance assigned to the various forces of evolution, three perspectives provide evolutionary explanations for molecular evolution.[18][19]Selectionist hypotheses argue that selection is the driving force of molecular evolution. While acknowledging that many mutations are neutral, selectionists attribute changes in the frequencies of neutral alleles to linkage disequilibrium with other loci that are under selection, rather than to random genetic drift.[20] Biases in codon usage are usually explained with reference to the ability of even weak selection to shape molecular evolution.[21]
Neutralist hypotheses emphasize the importance of mutation, purifying selection, and random genetic drift.[22] The introduction of the neutral theory by Kimura,[23] quickly followed by King and Jukes' own findings,[24] led to a fierce debate about the relevance of neodarwinism at the molecular level. The Neutral theory of molecular evolution proposes that most mutations in DNA are at locations not important to function or fitness. These neutral changes drift towards fixation within a population. Positive changes will be very rare, and so will not greatly contribute to DNA polymorphisms.[25] Deleterious mutations do not contribute much to DNA diversity because they negatively affect fitness and so are removed from the gene pool before long.[26] This theory provides a framework for the molecular clock.[25] The fate of neutral mutations are governed by genetic drift, and contribute to both nucleotide polymorphism and fixed differences between species.[27][28]
In the strictest sense, the neutral theory is not accurate.[29] Subtle changes in DNA very often have effects, but sometimes these effects are too small for natural selection to act on.[29] Even synonymous mutations are not necessarily neutral [29] because there is not a uniform amount of each codon. The nearly neutral theory expanded the neutralist perspective, suggesting that several mutations are nearly neutral, which means both random drift and natural selection is relevant to their dynamics.[29] The main difference between the neutral theory and nearly neutral theory is that the latter focuses on weak selection, not strictly neutral.[26]
Mutationists hypotheses emphasize random drift and biases in mutation patterns.[30] Sueoka was the first to propose a modern mutationist view. He proposed that the variation in GC content was not the result of positive selection, but a consequence of the GC mutational pressure.[31]
Protein evolution
This chart compares the sequence identity of different lipase
proteins
throughout the human body. It demonstrates how
proteins evolve, keeping
some regions conserved while others
change dramatically.
Evolution of proteins is studied by comparing the sequences and structures of proteins from many organisms representing distinct evolutionary clades. If the sequences/structures of two proteins are similar indicating that the proteins diverged from a common origin, these proteins are called as homologous proteins. More specifically, homologous proteins that exist in two distinct species are called as orthologs. Whereas, homologous proteins encoded by the genome of a single species are called paralogs.
The phylogenetic relationships of proteins are examined by multiple sequence comparisons. Phylogenetic trees of proteins can be established by the comparison of sequence identities among protoeins. Such phylogenetic trees have established that the sequence similarities among proteins reflect closely the evolutionary relationships among organisms.[32][33]
Protein evolution describes the changes over time in protein shape, function, and composition. Through quantitative analysis and experimentation, scientists have strived to understand the rate and causes of protein evolution. Using the amino acid sequences of hemoglobin and cytochrome c from multiple species, scientists were able to derive estimations of protein evolution rates. What they found was that the rates were not the same among proteins.[26] Each protein has its own rate, and that rate is constant across phylogenies (i.e., hemoglobin does not evolve at the same rate as cytochrome c, but hemoglobins from humans, mice, etc. do have comparable rates of evolution.). Not all regions within a protein mutate at the same rate; functionally important areas mutate more slowly and amino acid substitutions involving similar amino acids occurs more often than dissimilar substitutions.[26] Overall, the level of polymorphisms in proteins seems to be fairly constant. Several species (including humans, fruit flies, and mice) have similar levels of protein polymorphism.[25]