A Clovis projectile point created using bifacial percussion flaking (that is, each face is flaked on both edges alternately with a percussor)
The Clovis culture is a prehistoric Paleo-Indian culture, named for distinct stone tools found in close association with Pleistocene fauna at Blackwater Locality No. 1 near Clovis, New Mexico, in the 1920s and 1930s. It appears around 11,500–11,000 uncalibrated radiocarbon years before present at the end of the last glacial period, and is characterized by the manufacture of "Clovis points" and distinctive bone and ivory tools. Archaeologists' most precise determinations at present suggest this radiocarbon age is equal to roughly 13,200 to 12,900 calendar years ago. Clovis people are considered to be the ancestors of most of the indigenous cultures of the Americas.
The only human burial that has been directly associated with tools from the Clovis culture included the remains of an infant boy named Anzick-1.[5][6] Researchers from the United States and Europe conducted paleogenetic research on Anzick-1's ancient nuclear, mitochondrial, and Y-chromosome DNA.[7] The results of these analyses reveal that Anzick-1 is closely related to modern Native American populations, which lends support to the Beringia hypothesis for the settlement of the Americas.[8]
The Clovis culture was replaced by several more localized regional societies from the Younger Dryas cold-climate period onward. Post-Clovis cultures include the Folsom tradition, Gainey, Suwannee-Simpson, Plainview-Goshen, Cumberland,and Redstone. Each of these is thought to derive directly from Clovis, in some cases apparently differing only in the length of the fluting on their projectile points. Although this is generally held to be the result of normal cultural change through time,[9] numerous other reasons have been suggested as driving forces to explain changes in the archaeological record, such as the Younger Dryas postglacial climate change which exhibited numerous faunal extinctions.
After the discovery of several Clovis sites in eastern North America in the 1930s, the Clovis people came to be regarded as the first human inhabitants who created a widespread culture in the New World. However, this theory has been challenged, in the opinion of many archaeologists, by several archaeological discoveries, including sites such as Cactus Hill in Virginia, Paisley Caves in the Summer Lake Basin of Oregon, the Topper site in Allendale County, South Carolina, Meadowcroft Rockshelter in Pennsylvania, the Friedkin[10] site in Texas, Cueva Fell in Chile and, especially, Monte Verde, also in Chile.[11] The oldest claimed human archaeological site in the Americas is the Pedra Furada hearths, a site in Brazil that precedes the Clovis culture and the other sites already mentioned by 19,000 to 30,000 years. This claim has become an issue of contention between North American archaeologists and their South American and European counterparts, who disagree on whether it is conclusively proven to be an older human site.[12][13][14]
Description
Clovis points from the Rummells-Maske Cache Site, Iowa
A hallmark of the toolkit associated with the Clovis culture is the distinctively shaped, fluted stone spear point, known as the Clovis point. The Clovis point is bifacial and typically fluted on both sides. Archaeologists do not agree on whether the widespread presence of these artifacts indicates the proliferation of a single people, or the adoption of a superior technology by diverse population groups.[15]
The culture is named after artifacts found between 1932 and 1936 at Blackwater Locality No. 1, an archaeological site between the towns of Clovis and Portales, New Mexico. These finds were deemed especially important due to their direct association with mammoth sp. and the extinct Bison antiquus. The in situ finds of 1936 and 1937 included most of four stone Clovis points, two long bone points with impact damage, stone blades, a portion of a Clovis blade core, and several cutting tools made on stone flakes.[15] Clovis sites have since been identified throughout much of the contiguous United States, as well as Mexico and Central America, and even into northern South America.[16]
Clovis people are generally accepted to have hunted mammoths as well as extinct bison, mastodon, gomphotheres, sloths, tapir, camelops, horse and other smaller animals. More than 125 species of plants and animals are known to have been used by Clovis people in the portion of the Western Hemisphere they inhabited.[17][18]
The oldest Clovis site in North America is believed to be El Fin del Mundo in northwestern Sonora, Mexico, discovered during a 2007 survey. It features occupation dating around 13,390 calibrated years BP.[19] In 2011, remains of gomphotheres were found; the evidence suggests that humans did, in fact, kill two of them there. Also, the Aubrey site in Denton County, Texas, produced an almost identical radiocarbon date.[20]
Disappearance of Clovis
The most commonly held perspective on the end of the Clovis culture is that a decline in the availability of megafauna, combined with an overall increase in a less mobile population, led to local differentiation of lithic and cultural traditions across the Americas.[9][21] After this time, Clovis-style fluted points were replaced by other fluted-point traditions (such as the Folsom culture) with an essentially uninterrupted sequence across North and Central America. An effectively continuous cultural adaptation proceeds from the Clovis period through the ensuing Middle and Late Paleoindian periods.[22]Whether the Clovis culture drove the mammoth, and other species, to extinction via overhunting – the so-called Pleistocene overkill hypothesis – is still an open, and controversial, question.[23] It has also been hypothesized that the Clovis culture had its decline in the wake of the Younger Dryas cold phase.[24] This 'cold shock', lasting roughly 1500 years, affected many parts of the world, including North America. This appears to have been triggered by a vast amount of meltwater – possibly from Lake Agassiz – emptying into the North Atlantic, disrupting the thermohaline circulation.[25]
The Younger Dryas impact hypothesis or Clovis comet hypothesis originally proposed that a large air burst or earth impact of a comet or comets from outer space initiated the Younger Dryas cold period about 12,900 BP calibrated (10,900 14C uncalibrated) years ago.[26][27][28] The hypothesis has been largely contradicted by research showing that most of the findings cannot be repeated by other scientists, and criticized because of misinterpretation of data and the lack of confirmatory evidence.
Discovery
A cowboy and former slave, George McJunkin, found an ancient bison (Bison antiquus, an extinct relative of the American bison) skeleton in 1908 after a flash flood.[33] The site was first excavated in 1926, near Folsom, New Mexico, under the direction of Harold Cook and Jesse Figgins. On August 29, 1927, they found the first in situ Folsom point with the extinct B. antiquus bones. This confirmation of a human presence in the Americas during the Pleistocene inspired many people to start looking for evidence of early humans.[34]In 1929, 19-year-old Ridgely Whiteman, who had been closely following the excavations in nearby Folsom in the newspaper, discovered the Clovis site near the Blackwater Draw in eastern New Mexico. Despite several earlier Paleoindian discoveries, the best documented evidence of the Clovis complex was collected and excavated between 1932 and 1937 near Clovis, New Mexico, by a crew under the direction of Edgar Billings Howard until 1935 and later by John Cotter from the Academy of Natural Sciences/University of Pennsylvania. Howard's crew left their excavation in Burnet Cave, New Mexico, (the first truly professionally excavated Clovis site) in August, 1932, and visited Whiteman and his Blackwater Draw site. By November, Howard was back at Blackwater Draw to investigate additional finds from a construction project.[33]
The American Journal of Archaeology (January–March, 1932 V36 #1) in its "Archaeological Notes" mentions E. B. Howard's work in Burnet Cave, including the discovery of extinct fauna and a "Folsom type" point 4 ft below a Basketmaker burial. This brief mention of the Clovis point found in place antedates any work at the Dent Site in Colorado. Reference is made to a slightly earlier article on Burnet Cave in The University Museum Bulletin of November, 1931.
The first report of professional work at the Blackwater Draw Clovis site is in the November 25, 1932, issue of Science News. The publications on Burnet Cave and Blackwater Draw directly contradict statements by several authors (for example see Haynes 2002:56 The Early Settlement of North America) that Dent, Colorado was the first excavated Clovis site. The Dent Site, in Weld County, Colorado, was simply a fossil mammoth excavation in 1932. The first Dent Clovis point was found November 5, 1932, and the in situ point was found July 7, 1933. The in situ Clovis point from Burnet Cave was excavated in late August, 1931 (and reported early in 1932). E. B. Howard brought the Burnet Cave point to the 3rd Pecos Conference, September 1931, and showed it around to several archaeologists interested in early humans (see Woodbury 1983).
Also in 1968, in Montana, a Clovis burial site was found where the remains of a two-year-old child were studied. These remains have been named as Anzick-1 and recently, in 2014, have been used in scientific research.[7]
Clovis Paleo-Indians
Available genetic data show that the Clovis people are the direct ancestors of roughly 80% of all living Native American populations in North and South America, with the remainder descended from ancestors who entered in later waves of migration.[35][36] As reported in February 2014, DNA from the 12,600-year-old remains of Anzick boy, found in Montana, has affirmed this connection to the peoples of the Americas. In addition, this DNA analysis affirmed genetic connections back to ancestral peoples of northeast Asia. This adds weight to the theory that peoples migrated across a land bridge from Siberia to North America.[4]Clovis First/Single origin hypothesis
Known as "Clovis First", the predominant hypothesis among archaeologists in the latter half of the 20th century had been that the people associated with the Clovis culture were the first inhabitants of the Americas. The primary support for this was that no solid evidence of pre-Clovis human habitation had been found. According to the standard accepted theory, the Clovis people crossed the Beringia land bridge over the Bering Strait from Siberia to Alaska during the period of lowered sea levels during the ice age, then made their way southward through an ice-free corridor east of the Rocky Mountains in present-day Western Canada as the glaciers retreated.[37]This hypothesis came to be challenged by studies suggesting a pre-Clovis human occupation of the Americas.[38] In 2011, following the excavation of an occupation site at Buttermilk Creek, Texas, a prominent group of scientists claimed to have definitely established the existence "of an occupation older than Clovis."[39][40]
According to researchers Michael Waters and Thomas Stafford of Texas A&M University, new radiocarbon dates place Clovis remains from the continental United States in a shorter time window beginning 450 years later than the previously accepted threshold (13,200 to 12,900 BP).[2]
Recently, the scientific consensus has changed to acknowledge the presence of pre-Clovis cultures in the Americas, ending the "Clovis first" consensus.[41][42][43]
The results of a multiple-author study by Danish, Canadian, and American scientists published in Nature in February 2016 revealed that "the first Americans, whether Clovis or earlier groups in unglaciated North America before 12.6 cal. kyr BP", are "unlikely" to "have travelled to North America from Siberia via the Bering land bridge[44] "via a corridor that opened up between the melting ice sheets in what is now Alberta and B.C. about 13,000 years ago" as many anthropologists have argued for decades.[45] The lead author, Mikkel Pedersen – a PhD student from University of Copenhagen – explained, "The ice-free corridor was long considered the principal entry route for the first Americans ... Our results reveal that it simply opened up too late for that to have been possible."[45] The scientists argued that by 10,000 years ago, the ice-free corridor in what is now Alberta and B.C "was gradually taken over by a boreal forest dominated by spruce and pine trees" and that "Clovis people likely came from the south, not the north, perhaps following wild animals such as bison."[44][45]
Alternatives to Clovis-first
Evidence of human habitation before Clovis
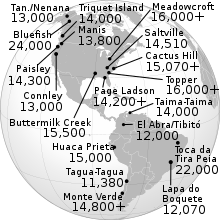
Archaeological sites that antedate Clovis that are well documented include:- Bluefish Caves, Yukon, Canada (24,000 yr BP)[46]
- Pedra Furada, Piauí, Brazil (10,500–12,000 yr BP; possibly >50,000 yr BP but this is disputed)[47][48]
- Topper, South Carolina, US (16,000–20,000 yr BP; possibly 50,000 yr BP but this is disputed)[49][50][51]
- Meadowcroft Rockshelter, Pennsylvania, US (16,000 yr BP)[52]
- Buttermilk Creek Complex, Salado, Texas, US (15,500 14C yr BP)[10][53][54]
- Cactus Hill, Virginia, US (15,070 14C yr BP)[55]
- Monte Verde, Chile (18,500 to 14,800[56] 14C yr BP)[57][58]
- Saltville (archaeological site), Virginia, US (14,510 14C yr BP)[59]
- Taima-Taima, Venezuela (14,000 yr BP)[60]
- Manis Mastodon Site, Sequim, Washington, US (13,800 yr BP)[61]
- Connley Caves, Oregon, US (13,000 yr BP)[62]
- Page-Ladson, Florida, US (14,550 cal yr BP])[63]
- Lapa do Boquete, Brazil (12,070 ±170 14C yr BP)[57][64]
- Paisley Caves, Oregon, US (14,300 cal yr BP)[65]
- Tanana Valley, Alaska, US (13,000–14,000 cal yr BP)[66]
- El Abra, Colombia (12,460 ±140 14C yr BP)[57]
- Nenana Valley, Alaska, US (12,000 yr BP)[67]
- Tibitó, Colombia (11,740 ±110 14C yr BP)[57]
- Tagua-Tagua, Chile (11,380 ±380 14C yr BP)[68]
Map of the Americas showing pre-Clovis sites
Predecessors of the Clovis people may have migrated south along the North American coastlines, although arguments exist for many migrations along several different routes.[69] Radiocarbon dating of the Monte Verde site in Chile places Clovis-like culture there as early as 18,500 to 14,500 years ago.[56] Remains found at the Channel Islands of California place coastal Paleoindians there 12,500 years ago. This suggests that the Paleoindian migration could have spread more quickly along the Pacific coastline, proceeding south, and that populations that settled along that route could have then begun migrations eastward into the continent.
The Pedra Furada sites in Brazil include a collection of rock shelters, which were used for thousands of years by diverse human populations. The first excavations yielded artifacts with carbon-14 dates of 48,000 to 32,000 years BP. Repeated analyses have confirmed this dating, carrying the range of dates up to 60,000 BP.[70] The best-analyzed archaeological levels are dated between 32,160 ± 1000 years BP and 17,000 ± 400 BP.
In 2004, worked stone tools were found at Topper in South Carolina that have been dated by radiocarbon techniques possibly to 50,000 years ago.[71] But, there is significant scholarly dispute regarding these dates.[72] Scholars agree that evidence of humans at the Topper Site date back to 22,900 cal yr BP.[73]
A more substantiated claim is that of Paisley Caves, Oregon, where rigorous carbon-14 and genetic testing appear to indicate that humans related to modern Native Americans were present in the caves over 1000 14C years before the earliest evidence of Clovis.[74] Traces and tools made by another people, the "Western Stemmed" tradition, were documented.[75]
A study published in Science presents strong evidence that humans occupied sites in Monte Verde, Chile, at the tip of South America, as early as 13,000 years ago.[76] If this is true, then humans must have entered North America long before the Clovis culture – perhaps 16,000 years ago.
The Tlapacoya site in Mexico is located along the base of a volcanic (remnant) hill on the shore of the former Lake Chalco. Seventeen excavations along the base of Tlapacoya Hill between 1956 and 1973 uncovered piles of disarticulated bones of bear and deer that appeared to have been butchered, plus 2,500 flakes and blades presumably from the butchering activities, plus one unfluted spear point. All were found in the same stratum containing three circular hearths filled with charcoal and ash. Bones of many other animal species were also present, including horses and migratory waterfowl. Two uncalibrated radiocarbon dates on carbon from the hearths came in around 24,000 and 22,000 years ago.[77] At another location, a prismatic microblade of obsidian was found in association with a tree trunk radiocarbon dated (uncalibrated) at roughly 24,000 years ago. This obsidian blade has recently been hydration dated by Joaquín García-Bárcena to 22,000 years ago. The hydration results were published in a seminal article that deals with the evidence for pre-Clovis habitation of Mexico.[78]
Coastal migration route
Studies of the mitochondrial DNA of First Nations/Native Americans published in 2007 suggest that the people of the New World may have diverged genetically from Siberians as early as 20,000 years ago, far earlier than the standard theory suggests.[38] According to one alternative theory, the Pacific coast of North America may have been free of ice, allowing the first peoples in North America to come down this route prior to the formation of the ice-free corridor in the continental interior.[79] No evidence has yet been found to support this hypothesis[citation needed] except that genetic analysis of coastal marine life indicates diverse fauna persisting in refugia throughout the Pleistocene ice ages along the coasts of Alaska and British Columbia; these refugia include common food sources of coastal aboriginal peoples, suggesting that a migration along the coastline was feasible at the time.[80] Some early sites on the coast, for example Namu, British Columbia, exhibit maritime focus on foods from an early point with substantial cultural continuity.[81]In February 2014, researchers reported on their DNA analysis of the remains of Anzick boy (referred to as Anzick-1) of Montana, the oldest skeleton found in the Americas and dated to 12,600 years ago. They found the mtDNA to be D4h3a, "one of the rare lineages associated with Native Americans."[7] This was the same as the mtDNA associated with current coastal populations in North and South America. The study team suggest that finding this genetic evidence so far inland shows that "current distribution of genetic markers are not necessarily indicative of the movement or distribution of peoples in the past."[7] The Y haplotype was found to be Q-L54*(xM3). Further testing found that Anzick-1 was most closely related to Native American populations (see below).[7]
Solutrean hypothesis
The controversial Solutrean hypothesis proposed in 1999 by Smithsonian archaeologist Dennis Stanford and colleague Bruce Bradley (Stanford and Bradley 2002), suggests that the Clovis people could have inherited technology from the Solutrean people who lived in southern Europe 21,000–15,000 years ago, and who created the first Stone Age artwork in present-day southern France.[82] The link is suggested by the similarity in technology between the projectile points of the Solutreans and those found at Clovis (and pre-Clovis) sites. Its proponents point to tools found at various pre-Clovis sites in eastern North America (particularly in the Chesapeake Bay region) as progenitors of Clovis-style tools.[83] The model envisions these people making the crossing in small watercraft via the edge of the pack ice in the North Atlantic Ocean that then extended to the Atlantic coast of France, using skills similar to those of the modern Inuit people, making landfall somewhere around the then-exposed Grand Banks of the North American continental shelf.In a 2008 study of the relevant paleoceanographic data, Kieran Westley and Justin Dix concluded that "it is clear from the paleoceanographic and paleo-environmental data that the Last Glacial Maximum (LGM) North Atlantic does not fit the descriptions provided by the proponents of the Solutrean Atlantic Hypothesis. Although ice use and sea mammal hunting may have been important in other contexts, in this instance, the conditions militate against an ice-edge-following, maritime-adapted European population reaching the Americas."[84]
University of New Mexico anthropologist Lawrence G. Straus, a primary critic of the Solutrean hypothesis, points to the theoretical difficulty of the ocean crossing, a lack of Solutrean-specific features in pre-Clovis artifacts, as well as the lack of art (such as that found at Lascaux in France) among the Clovis people, as major deficiencies in the Solutrean hypothesis. The 3,000 to 5,000 radiocarbon year gap between the Solutrean period of France and Spain and the Clovis of the New World also makes such a connection problematic.[85] In response, Bradley and Stanford contend that it was "a very specific subset of the Solutrean who formed the parent group that adapted to a maritime environment and eventually made it across the north Atlantic ice-front to colonize the east coast of the Americas" and that this group may not have shared all Solutrean cultural traits.[86]
Genetic evidence of east/west dichotomy
Mitochondrial DNA analysis in 2014 has found that members of some native North American tribes have a maternal ancestry (called haplogroup X) linked to the maternal ancestors of some present-day individuals in western Asia and Europe, albeit distantly. This has also provided some support for pre-Clovis models. More specifically, a variant of mitochondrial DNA called X2a found in many Native Americans has been traced to western Eurasia, while not being found in eastern Eurasia.[87]Mitochondrial DNA analysis of Anzick-1 concluded that the boy belonged to what is known as haplogroup or lineage D4h3a. This finding is important because the D4h3a line is considered to be a lineage "founder", belonging to the first people to reach the Americas. Although rare in most of today's Native Americans in the US and Canada, D4h3a genes are more common among native peoples of South America, far from the site in Montana where Anzick-1 was buried. This suggests a greater genetic complexity among Native Americans than previously thought, including an early divergence in the genetic lineage 13,000 years ago. One theory suggests that after crossing into North America from Siberia, a group of the first Americans, with the lineage D4h3a, moved south along the Pacific coast and, over thousands of years, into Central and South America, while others may have moved inland, east of the Rocky Mountains.[7] The apparent early divergence between North American and Central plus South American populations may or may not be associated with post-divergence gene flow from a more basal population into North America; however, analysis of published DNA sequences for 19 Siberian populations does not favor the latter scenario.[7]
Spearheads and DNA found at the Paisley Caves site in Oregon suggest that North America was colonized by more than one culture, and that the Clovis culture was not the first. There is evidence to suggest an east/west dichotomy, with the Clovis culture located to the east.[88]
But in 2014, the autosomal DNA of a 12,500+-year-old infant from Montana was sequenced.[7][8][89][90] The DNA was taken from a skeleton referred to as Anzick-1, found in close association with several Clovis artifacts. Comparisons indicate strong affinities with DNA from Siberian sites, and virtually rule out close affinity with European sources (the "Solutrean hypothesis"). The DNA shows strong affinities with all existing Native American populations, which indicated that each of them derives from an ancient population that lived in or near Siberia, the Upper Palaeolithic Mal'ta population. Mal'ta belonged to Y-DNA haplogroup R and mitochrondrial macrohaplogroup U.[7][91]
The data indicate that Anzick-1 is from a population directly ancestral to present South American and Central American Native American populations. This rules out hypotheses which posit that invasions subsequent to the Clovis culture overwhelmed or assimilated previous migrants into the Americas. Anzick-1 is less closely related to present North American Native American populations (including a Yaqui genetic sample), suggesting that the North American populations are basal to Anzick-1 and Central and South American populations.[7] The apparent early divergence between North American and Central plus South American populations might be due to post-divergence gene flow from a more basal population into North America; however, analysis of published DNA sequences of 19 Siberian populations do not suggest this scenario.[7] Anzick-1 belonged to Y-haplogroup Q-L54(xM3)[7], which is by far the largest haplogroup among Native Americans.
Other sites
Mammuthus primigenius "Hebior Mammoth specimen" bearing tool/butcher marks, cast skeleton produced and distributed by Triebold Paleontology Incorporated
In approximate reverse chronological order:
- Pedra Furada, Serra da Capivara National Park, in the state of Piauí, Brazil. Site with evidence of non-Clovis human remains, a rock painting rupestre art drawings from at least 12,000–6,000 BP. Hearth samples C-14 dates of 48–32,000 BP were reported in a Nature article (Guidon and Delibrias 1986). New hearth samples with ABOX dates of 54,000 BP were reported in the Quaternary Science Reviews.[92] Paleoindian components found here, have been challenged by American researchers such as Meltzer, Adovasio, and Dillehay.
- The Monte Verde site in Chile, was occupied from 14,800 years BP,[93] with bones and other finds dating on average 12,500 yrs BP.[94] The earliest finds at the site were from between 32,840 and 33,900 years BP, but are controversial.[94]
- The Bluefish Caves site in Yukon, Canada, contains bones with evidence of human cut-marks which demonstrates a human presence as early as 24,000 yr BP. The Bluefish caves are currently the oldest archaeological site in North America and offers evidence regarding the Beringia Standstill hypothesis, which states a genetically isolated human population remained in the area during the last glacial maximum and then traveled within North America and South America after the glaciers receded.[95]
- Lagoa Santa, Minas Gerais, Brazil, is erroneously asserted to be Clovis age or even possibly Pre-Clovis in age. The recent discussion of this site (specifically Lapa Vermelha IV) and the Luzia skull, reportedly 11,500 years old by Neves and Hubb, makes it clear that this date is a chronological date in years Before Present and not a raw radiocarbon date[96] in eastern Brazil. Clovis sites mostly date between 11,500 and 11,000 radiocarbon years which means 13,000 years before present at a minimum. "Luzia" is at least 1,000 years younger than Clovis and Lapa Vermelha IV should not be considered a Pre-Clovis site.
- Cueva del Milodón, in Patagonian Chile[97] dates at least as early as 10,500 BP. This is a site found particularly early in the New World hunt for Early Man, circa 1896, and needs additional basic research, but 10,500 B.P. would be 1,500 years younger than Clovis, or if the dating is 10,500 RCYBP, it would still be roughly 500–700 years younger than Clovis. In either case this should not be considered a Pre-Clovis site.
- Cueva Fell[98] and Pali Aike Crater sites in Patagonia, with hearths, stone tools and other elements of human habitation dating to at least as early as 11,000 BP.
- The Big Eddy Site in southwestern Missouri contains several claimed pre-Clovis artifacts or geofacts. In situ artifacts have been found in this well-stratified site in association with charcoal. Five different samples have been AMS dated to between 11,300 and 12,675 BP (Before Present).[citation needed]
- Taima Taima, Venezuela has cultural material very similar to Monte Verde II, dating to 12,000 years BP.[citation needed] Recovered artifacts of the El Jobo complex in direct association with the butchered remains of a juvenile mastodon. Radiocarbon dates on associated wood twigs indicate a minimum age of 13,000 years before the present for the mastodon kill, a dating significantly older than that of the Clovis complex in North America.[99]
- A cut mastodon tusk found at Page-Ladson, Jefferson County, Florida on the Aucilla River has been dated to 12,300 years BP near a few in situ artifacts of similar age.[100]
- The Schaefer and Hebior mammoth sites in Kenosha County, Wisconsin indicate exploitation of this animal by humans. The Schaefer Mammoth site has over 13 highly purified collagen AMS dates and 17 dates on associated wood, dating it to 12,300–12,500 radiocarbon years before the present. Hebior has two AMS dates in the same range. Both animals show conclusive butchering marks and associated non-diagnostic tools.[101]
- A site in Walker, Minnesota with stone tools, alleged to be from 13,000 to 15,000 years old based on surrounding geology, was discovered in 2006.[102] However, further examination suggests that the site does not represent a human occupation.[103]
- In a 2011 article[10] in Science, Waters et al. 2011 describe an assemblage of 15,528 lithic artifacts from the Debra L. Friedkin site west of Salado, Texas. These artifacts (including 56 tools, 2,268 macrodebitage and 13,204 microdebitage) define the Buttermilk Creek Complex formation, which stratigraphically underlies a Clovis assemblage. While carbon dating could not be used to directly date the artifacts, 49 samples from the 20 cm Buttermilk floodplain sedimentary clay layer in which the artifacts were embedded were dated using optically stimulated luminescence (OSL). Eighteen OSL ages, ranging from 14,000 to 17,500 ka were obtained from this layer. The authors report "the most conservative estimate" of the age of the Buttermilk clays range from 13,200 to 15,500 ka, based on the minimum age represented by each of the 18 OSL ages.
- Human coprolites have been found in Paisley Caves in Oregon, carbon dated at 14,300 years ago. Genetic analysis revealed that the coprolites contained mtDNA haplogroups A2 and B2, two of the five major Native American mtDNA haplogroups.[104][105]
- The Mud Lake site, in Kenosha County, Wisconsin consists of the foreleg of a juvenile mammoth recovered in the 1930s. Over 100 stone tool butchering marks are found on the bones. Several purified collagen AMS dates show the animal to be 13,450 RCYBP with a range of plus or minus 1,500 RCYBP variance.[106]
- Meadowcroft Rockshelter in southwestern Pennsylvania, excavated 1973–78, with evidence of occupancy dating back from 16,000 to 19,000 years ago.[107]
- Cactus Hill in southern Virginia, with artifacts such as unfluted bifacial stone tools with dates ranging from c. 15,000 to 17,000 years ago.[108]
- Sixty-eight stone and bone tools discovered in an orchard in East Wenatchee, Washington in 1987, excavated in 1988 and 1990. Five of the Clovis points are on display at the Wenatchee Valley Museum & Cultural Center.