The history of biochemistry can be said to have started with the ancient Greeks who were interested in the composition and processes of life, although biochemistry as a specific scientific discipline has its beginning around the early 19th century. Some argued that the beginning of biochemistry may have been the discovery of the first enzyme, diastase (today called amylase), in 1833 by Anselme Payen, while others considered Eduard Buchner's first demonstration of a complex biochemical process alcoholic fermentation in cell-free extracts to be the birth of biochemistry. Some might also point to the influential work of Justus von Liebig from 1842, Animal chemistry, or, Organic chemistry in its applications to physiology and pathology, which presented a chemical theory of metabolism, or even earlier to the 18th century studies on fermentation and respiration by Antoine Lavoisier.
The term “biochemistry” itself is derived from the combining form bio-, meaning "life", and chemistry. The word is first recorded in English in 1848, while in 1877, Felix Hoppe-Seyler used the term (Biochemie in German) in the foreword to the first issue of Zeitschrift für Physiologische Chemie (Journal of Physiological Chemistry) as a synonym for physiological chemistry and argued for the setting up of institutes dedicate to its studies. Nevertheless, several sources cite German chemist Carl Neuberg as having coined the term for the new discipline in 1903, and some credit it to Franz Hofmeister.
The subject of study in biochemistry is the chemical processes in living organisms, and its history involves the discovery and understanding of the complex components of life and the elucidation of pathways of biochemical processes. Much of biochemistry deals with the structures and functions of cellular components such as proteins, carbohydrates, lipids, nucleic acids and other biomolecules; their metabolic pathways and flow of chemical energy through metabolism; how biological molecules give rise to the processes that occur within living cells; it also focuses on the biochemical processes involved in the control of information flow through biochemical signalling, and how they relate to the functioning of whole organisms. Over the last 40 years the field has had success in explaining living processes such that now almost all areas of the life sciences from botany to medicine are engaged in biochemical research.
Among the vast number of different biomolecules, many are complex and large molecules (called polymers), which are composed of similar repeating subunits (called monomers). Each class of polymeric biomolecule has a different set of subunit types. For example, a protein is a polymer whose subunits are selected from a set of twenty or more amino acids, carbohydrates are formed from sugars known as monosaccharides, oligosaccharides, and polysaccharides, lipids are formed from fatty acids and glycerols, and nucleic acids are formed from nucleotides. Biochemistry studies the chemical properties of important biological molecules, like proteins, and in particular the chemistry of enzyme-catalyzed reactions. The biochemistry of cell metabolism and the endocrine system has been extensively described. Other areas of biochemistry include the genetic code (DNA, RNA), protein synthesis, cell membrane transport, and signal transduction.
Proto-biochemistry
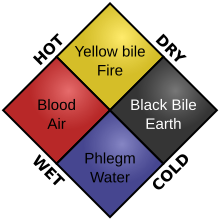
The
four humors. In this diagram, each kind of food would result in a
different physiological result. For example, cold and dry food would
produce black bile.
In a sense, the study of biochemistry can be considered to have started in ancient times, for example when biology first began to interest society—as the ancient Chinese developed a system of medicine based on yin and yang, and also the five phases,
which both resulted from alchemical and biological interests. Its
beginning in the ancient Indian culture was linked to an interest in
medicine, as they developed the concept of three humors that were
similar to the Greek's four humours. They also delved into the interest of bodies being composed of tissues.
The ancient Greeks conception of "biochemistry" was linked with their
ideas on matter and disease, where good health was thought to come from a
balance of the four elements and four humors in the human body.
As in the majority of early sciences, the Islamic world contributed
significantly to early biological advancements as well as alchemical
advancements; especially with the introduction of clinical trials and clinical pharmacology presented in Avicenna's The Canon of Medicine. On the side of chemistry, early advancements were heavily attributed to exploration of alchemical interests but also included: metallurgy, the scientific method, and early theories of atomism. In more recent times, the study of chemistry was marked by milestones such as the development of Mendeleev's periodic table, Dalton's atomic model, and the conservation of mass theory. This last mention has the most importance of the three due to the fact that this law intertwines chemistry with thermodynamics in an intercalated manner.
Enzymes
As early as the late 18th century and early 19th century, the digestion of meat by stomach secretions and the conversion of starch to sugars by plant extracts and saliva were known. However, the mechanism by which this occurred had not been identified.
In the 19th century, when studying the fermentation of sugar to alcohol by yeast, Louis Pasteur concluded that this fermentation was catalyzed by a vital force contained within the yeast cells called ferments,
which he thought functioned only within living organisms. He wrote that
"alcoholic fermentation is an act correlated with the life and
organization of the yeast cells, not with the death or putrefaction of
the cells."
Anselme Payen discovered in 1833 the first enzyme who called diastase and in 1878 German physiologist Wilhelm Kühne (1837–1900) coined the term enzyme, which comes from Greek ενζυμον "in leaven", to describe this process. The word enzyme was used later to refer to nonliving substances such as pepsin, and the word ferment used to refer to chemical activity produced by living organisms.
In 1897 Eduard Buchner
began to study the ability of yeast extracts to ferment sugar despite
the absence of living yeast cells. In a series of experiments at the University of Berlin, he found that the sugar was fermented even when there were no living yeast cells in the mixture. He named the enzyme that brought about the fermentation of sucrose "zymase". In 1907 he received the Nobel Prize in Chemistry
"for his biochemical research and his discovery of cell-free
fermentation". Following Buchner's example; enzymes are usually named
according to the reaction they carry out. Typically the suffix -ase is added to the name of the substrate (e.g., lactase is the enzyme that cleaves lactose) or the type of reaction (e.g., DNA polymerase forms DNA polymers).
Shown above is the restriction endonuclease EcoR1 in its 3D computer generated form.
Having shown that enzymes could function outside a living cell, the
next step was to determine their biochemical nature. Many early workers
noted that enzymatic activity was associated with proteins, but several
scientists (such as Nobel laureate Richard Willstätter) argued that proteins were merely carriers for the true enzymes and that proteins per se were incapable of catalysis. However, in 1926, James B. Sumner showed that the enzyme urease was a pure protein and crystallized it; Sumner did likewise for the enzyme catalase in 1937. The conclusion that pure proteins can be enzymes was definitively proved by Northrop and Stanley,
who worked on the digestive enzymes pepsin (1930), trypsin and
chymotrypsin. These three scientists were awarded the 1946 Nobel Prize
in Chemistry.
This discovery, that enzymes could be crystallized, meant that scientists eventually could solve their structures by x-ray crystallography. This was first done for lysozyme, an enzyme found in tears, saliva and egg whites that digests the coating of some bacteria; the structure was solved by a group led by David Chilton Phillips and published in 1965. This high-resolution structure of lysozyme marked the beginning of the field of structural biology and the effort to understand how enzymes work at an atomic level of detail.
Metabolism
Early metabolic interest
Santorio Santorio in his steelyard balance, from Ars de statica medecina, first published 1614
The term metabolism is derived from the Greek Μεταβολισμός – Metabolismos for "change", or "overthrow".
The history of the scientific study of metabolism spans 800 years. The
earliest of all metabolic studies began during the early thirteenth
century (1213-1288) by a Muslim scholar from Damascus named Ibn
al-Nafis. al-Nafis stated in his most well-known work Theologus Autodidactus
that "that body and all its parts are in a continuous state of
dissolution and nourishment, so they are inevitably undergoing permanent
change."
Although al-Nafis was the first documented physician to have an
interest in biochemical concepts, the first controlled experiments in
human metabolism were published by Santorio Santorio in 1614 in his book Ars de statica medecina.
This book describes how he weighed himself before and after eating,
sleeping, working, sex, fasting, drinking, and excreting. He found that
most of the food he took in was lost through what he called "insensible
perspiration".
Metabolism: 20th century - present
One of the most prolific of these modern biochemists was Hans Krebs who made huge contributions to the study of metabolism.
Krebs was a student of extremely important Otto Warburg, and wrote a
biography of Warburg by that title in which he presents Warburg as being
educated to do for biological chemistry what Fischer did for organic
chemistry. Which he did. Krebs discovered the urea cycle and later,
working with Hans Kornberg, the citric acid cycle and the glyoxylate cycle. These discoveries led to Krebs being awarded the Nobel Prize in physiology in 1953, which was shared with the German biochemist Fritz Albert Lipmann who also codiscovered the essential cofactor coenzyme A.
Glucose absorption
In 1960, the biochemist Robert K. Crane revealed his discovery of the sodium-glucose cotransport as the mechanism for intestinal glucose absorption.
This was the very first proposal of a coupling between the fluxes of
an ion and a substrate that has been seen as sparking a revolution in
biology. This discovery, however, would not have been possible if it
were not for the discovery of the molecule glucose's structure and chemical makeup. These discoveries are largely attributed to the German chemist Emil Fischer who received the Nobel Prize in chemistry nearly 60 years earlier.
Glycolysis
Shown here is a step-wise depiction of glycolysis along with the required enzymes.
Since metabolism focuses on the breaking down (catabolic processes)
of molecules and the building of larger molecules from these particles
(anabolic processes), the use of glucose and its involvement in the
formation of adenosine triphosphate (ATP) is fundamental to this understanding. The most frequent type of glycolysis found in the body is the type that follows the Embden-Meyerhof-Parnas (EMP) Pathway, which was discovered by Gustav Embden, Otto Meyerhof, and Jakob Karol Parnas.
These three men discovered that glycolysis is a strongly determinant
process for the efficiency and production of the human body. The
significance of the pathway shown in the adjacent image is that by
identifying the individual steps in this process doctors and researchers
are able to pinpoint sites of metabolic malfunctions such as pyruvate kinase deficiency
that can lead to severe anemia. This is most important because cells,
and therefore organisms, are not capable of surviving without proper
functioning metabolic pathways.
Instrumental advancements (20th century)
Since then, biochemistry has advanced, especially since the mid-20th century, with the development of new techniques such as chromatography, X-ray diffraction, NMR spectroscopy, radioisotopic labelling, electron microscopy and molecular dynamics simulations. These techniques allowed for the discovery and detailed analysis of many molecules and metabolic pathways of the cell, such as glycolysis and the Krebs cycle (citric acid cycle). The example of an NMR instrument shows that some of these instruments, such as the HWB-NMR,
can be very large in size and can cost anywhere from a few hundred
dollars to millions of dollars ($16 million for the one shown here).
Polymerase chain reaction
Shown above is a model of a thermo cycler that is currently being used in polymerase chain reaction.
Polymerase chain reaction (PCR) is the primary gene amplification
technique that has revolutionized modern biochemistry. Polymerase chain
reaction was developed by Kary Mullis in 1983.
There are four steps to a proper polymerase chain reaction: 1)
denaturation 2) extension 3) insertion (of gene to be expressed) and
finally 4) amplification of the inserted gene. These steps with simple
illustrative examples of this process can be seen in the image below and
to the right of this section. This technique allows for the copy of a
single gene to be amplified into hundreds or even millions of copies and
has become a cornerstone in the protocol for any biochemist that wishes
to work with bacteria and gene expression. PCR is not only used for
gene expression research but is also capable of aiding laboratories in
diagnosing certain diseases such a lymphomas, some types of leukemia, and other malignant
diseases that can sometimes puzzle doctors. Without polymerase chain
reaction development, there are many advancements in the field of
bacterial study and protein expression study that would not have come to
fruition. The development of the theory and process of polymerase chain reaction is essential but the invention of the thermal cycler
is equally as important because the process would not be possible
without this instrument. This is yet another testament to the fact that
the advancement of technology is just as crucial to sciences such as
biochemistry as is the painstaking research that leads to the
development of theoretical concepts.