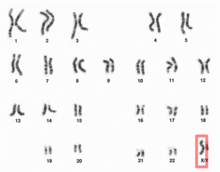
| Human Y chromosome | |
|---|---|

Human Y chromosome (after G-banding)
| |
Y chromosome in human male karyogram
| |
| Features | |
| Length (bp) | 57,227,415 bp (GRCh38) |
| No. of genes | 63 (CCDS) |
| Type | Allosome |
| Centromere position | Acrocentric (10.4 Mbp) |
| Complete gene lists | |
| CCDS | Gene list |
| HGNC | Gene list |
| UniProt | Gene list |
| NCBI | Gene list |
| External map viewers | |
| Ensembl | Chromosome Y |
| Entrez | Chromosome Y |
| NCBI | Chromosome Y |
| UCSC | Chromosome Y |
| Full DNA sequences | |
| RefSeq | NC_000024 (FASTA) |
| GenBank | CM000686 (FASTA) |
The Y chromosome is one of two sex chromosomes (allosomes) in mammals, including humans, and many other animals. The other is the X chromosome. Y is normally the sex-determining chromosome in many species, since it is the presence or absence of Y that typically determines the male or female sex of offspring produced in sexual reproduction. In mammals, the Y chromosome contains the gene SRY, which by default triggers male development. The DNA in the human Y chromosome is composed of about 59 million base pairs. The Y chromosome is passed only from father to son. With a 30% difference between humans and chimpanzees, the Y chromosome is one of the fastest-evolving parts of the human genome. To date, over 200 Y-linked genes have been identified. All Y-linked genes are expressed and (apart from duplicated genes) hemizygous (present on only one chromosome) except in the cases of aneuploidy such as XYY syndrome or XXYY syndrome.
Overview
Discovery
The Y chromosome was identified as a sex-determining chromosome by Nettie Stevens at Bryn Mawr College in 1905 during a study of the mealworm Tenebrio molitor. Edmund Beecher Wilson
independently discovered the same mechanisms the same year. Stevens
proposed that chromosomes always existed in pairs and that the Y
chromosome was the pair of the X chromosome discovered in 1890 by Hermann Henking. She realized that the previous idea of Clarence Erwin McClung, that the X chromosome determines sex, was wrong and that sex determination
is, in fact, due to the presence or absence of the Y chromosome.
Stevens named the chromosome "Y" simply to follow on from Henking's "X"
alphabetically.
The idea that the Y chromosome was named after its similarity in
appearance to the letter "Y" is mistaken. All chromosomes normally
appear as an amorphous blob under the microscope and only take on a
well-defined shape during mitosis. This shape is vaguely X-shaped for all chromosomes. It is entirely coincidental that the Y chromosome, during mitosis, has two very short branches which can look merged under the microscope and appear as the descender of a Y-shape.
Variations
Most therian mammals have only one pair of sex chromosomes in each cell. Males have one Y chromosome and one X chromosome, while females have two X chromosomes. In mammals, the Y chromosome contains a gene, SRY,
which triggers embryonic development as a male. The Y chromosomes of
humans and other mammals also contain other genes needed for normal
sperm production.
There are exceptions, however. Among humans, some men have two Xs and a Y ("XXY", see Klinefelter syndrome), or one X and two Ys, and some women have three Xs or a single X instead of a double X. There are other exceptions in which SRY is damaged (leading to an XY female), or copied to the X (leading to an XX male).
Origins and evolution
Before Y chromosome
Many ectothermic vertebrates
have no sex chromosomes. If they have different sexes, sex is
determined environmentally rather than genetically. For some of them,
especially reptiles, sex depends on the incubation temperature; others are hermaphroditic (meaning they contain both male and female gametes in the same individual).
Origin
The X and Y chromosomes are thought to have evolved from a pair of identical chromosomes, termed autosomes, when an ancestral animal developed an allelic variation, a so-called "sex locus" – simply possessing this allele caused the organism to be male.
The chromosome with this allele became the Y chromosome, while the
other member of the pair became the X chromosome. Over time, genes that
were beneficial for males and harmful to (or had no effect on) females
either developed on the Y chromosome or were acquired through the
process of translocation.
Until recently, the X and Y chromosomes were thought to have diverged around 300 million years ago. However, research published in 2010, and particularly research published in 2008 documenting the sequencing of the platypus genome,
has suggested that the XY sex-determination system would not have been
present more than 166 million years ago, at the split of the monotremes from other mammals. This re-estimation of the age of the therian XY system is based on the finding that sequences that are on the X chromosomes of marsupials and eutherian mammals are present on the autosomes of platypus and birds. The older estimate was based on erroneous reports that the platypus X chromosomes contained these sequences.
Recombination inhibition
Recombination
between the X and Y chromosomes proved harmful—it resulted in males
without necessary genes formerly found on the Y chromosome, and females
with unnecessary or even harmful genes previously only found on the Y
chromosome. As a result, genes beneficial to males accumulated near the
sex-determining genes, and recombination in this region was suppressed
in order to preserve this male specific region.
Over time, the Y chromosome changed in such a way as to inhibit the
areas around the sex determining genes from recombining at all with the X
chromosome. As a result of this process, 95% of the human Y chromosome
is unable to recombine. Only the tips of the Y and X chromosomes
recombine. The tips of the Y chromosome that could recombine with the X
chromosome are referred to as the pseudoautosomal region. The rest of the Y chromosome is passed on to the next generation intact, allowing for its use in tracking human evolution.
Degeneration
By one estimate, the human Y chromosome has lost 1,393 of its 1,438 original genes over the course of its existence, and linear extrapolation of this 1,393-gene loss over 300 million years gives a rate of genetic loss of 4.6 genes per million years.
Continued loss of genes at the rate of 4.6 genes per million years
would result in a Y chromosome with no functional genes – that is the Y
chromosome would lose complete function – within the next 10 million
years, or half that time with the current age estimate of 160 million
years.
Comparative genomic analysis reveals that many mammalian species are
experiencing a similar loss of function in their heterozygous sex
chromosome. Degeneration may simply be the fate of all non-recombining
sex chromosomes, due to three common evolutionary forces: high mutation rate, inefficient selection, and genetic drift.
However, comparisons of the human and chimpanzee
Y chromosomes (first published in 2005) show that the human Y
chromosome has not lost any genes since the divergence of humans and
chimpanzees between 6–7 million years ago,
and a scientific report in 2012 stated that only one gene had been lost
since humans diverged from the rhesus macaque 25 million years ago.
These facts provide direct evidence that the linear extrapolation
model is flawed and suggest that the current human Y chromosome is
either no longer shrinking or is shrinking at a much slower rate than
the 4.6 genes per million years estimated by the linear extrapolation
model.
High mutation rate
The
human Y chromosome is particularly exposed to high mutation rates due
to the environment in which it is housed. The Y chromosome is passed
exclusively through sperm, which undergo multiple cell divisions during gametogenesis.
Each cellular division provides further opportunity to accumulate base
pair mutations. Additionally, sperm are stored in the highly oxidative
environment of the testis, which encourages further mutation. These two
conditions combined put the Y chromosome at a greater opportunity of
mutation than the rest of the genome. The increased mutation opportunity for the Y chromosome is reported by Graves as a factor 4.8.
However, her original reference obtains this number for the relative
mutation rates in male and female germ lines for the lineage leading to
humans.
The observation that the Y chromosome experiences little meiotic recombination and has an accelerated rate of mutation and degradative change compared to the rest of the genome suggests an evolutionary explanation for the adaptive function of meiosis with respect to the main body of genetic information. Brandeis
proposed that the basic function of meiosis (particularly meiotic
recombination) is the conservation of the integrity of the genome, a
proposal consistent with the idea that meiosis is an adaptation for repairing DNA damage.
Inefficient selection
Without the ability to recombine during meiosis, the Y chromosome is unable to expose individual alleles
to natural selection. Deleterious alleles are allowed to "hitchhike"
with beneficial neighbors, thus propagating maladapted alleles in to the
next generation. Conversely, advantageous alleles may be selected
against if they are surrounded by harmful alleles (background
selection). Due to this inability to sort through its gene content, the Y
chromosome is particularly prone to the accumulation of "junk" DNA. Massive accumulations of retrotransposable elements are scattered throughout the Y.
The random insertion of DNA segments often disrupts encoded gene
sequences and renders them nonfunctional. However, the Y chromosome has
no way of weeding out these "jumping genes". Without the ability to
isolate alleles, selection cannot effectively act upon them.
A clear, quantitative indication of this inefficiency is the entropy rate of the Y chromosome. Whereas all other chromosomes in the human genome
have entropy rates of 1.5–1.9 bits per nucleotide (compared to the
theoretical maximum of exactly 2 for no redundancy), the Y chromosome's
entropy rate is only 0.84. This means the Y chromosome has a much lower information content relative to its overall length; it is more redundant.
Genetic drift
Even
if a well adapted Y chromosome manages to maintain genetic activity by
avoiding mutation accumulation, there is no guarantee it will be passed
down to the next generation. The population size of the Y chromosome is
inherently limited to 1/4 that of autosomes: diploid organisms contain
two copies of autosomal chromosomes while only half the population
contains 1 Y chromosome. Thus, genetic drift is an exceptionally strong
force acting upon the Y chromosome. Through sheer random assortment, an
adult male may never pass on his Y chromosome if he only has female
offspring. Thus, although a male may have a well adapted Y chromosome
free of excessive mutation, it may never make it into the next gene
pool.
The repeat random loss of well-adapted Y chromosomes, coupled with the
tendency of the Y chromosome to evolve to have more deleterious
mutations rather than less for reasons described above, contributes to
the species-wide degeneration of Y chromosomes through Muller's ratchet.
Gene conversion
As it has been already mentioned, the Y chromosome is unable to recombine during meiosis like the other human chromosomes; however, in 2003, researchers from MIT discovered a process which may slow down the process of degradation.
They found that human Y chromosome is able to "recombine" with itself, using palindrome base pair sequences. Such a "recombination" is called gene conversion.
In the case of the Y chromosomes, the palindromes are not noncoding DNA;
these strings of bases contain functioning genes important for male
fertility. Most of the sequence pairs are greater than 99.97% identical.
The extensive use of gene conversion may play a role in the ability of
the Y chromosome to edit out genetic mistakes and maintain the integrity
of the relatively few genes it carries. In other words, since the Y
chromosome is single, it has duplicates of its genes on itself instead
of having a second, homologous, chromosome. When errors occur, it can
use other parts of itself as a template to correct them.
Findings were confirmed by comparing similar regions of the Y chromosome in humans to the Y chromosomes of chimpanzees, bonobos and gorillas.
The comparison demonstrated that the same phenomenon of gene conversion
appeared to be at work more than 5 million years ago, when humans and
the non-human primates diverged from each other.
Future evolution
In
the terminal stages of the degeneration of the Y chromosome, other
chromosomes increasingly take over genes and functions formerly
associated with it. Finally, the Y chromosome disappears entirely, and a
new sex-determining system arises. Several species of rodent in the sister families Muridae and Cricetidae have reached these stages, in the following ways:
- The Transcaucasian mole vole, Ellobius lutescens, the Zaisan mole vole, Ellobius tancrei, and the Japanese spinous country rats Tokudaia osimensis and Tokudaia tokunoshimensis, have lost the Y chromosome and SRY entirely. Tokudaia spp. have relocated some other genes ancestrally present on the Y chromosome to the X chromosome. Both sexes of Tokudaia spp. and Ellobius lutescens have an XO genotype (Turner syndrome), whereas all Ellobius tancrei possess an XX genotype. The new sex-determining system(s) for these rodents remains unclear.
- The wood lemming Myopus schisticolor, the Arctic lemming, Dicrostonyx torquatus, and multiple species in the grass mouse genus Akodon have evolved fertile females who possess the genotype generally coding for males, XY, in addition to the ancestral XX female, through a variety of modifications to the X and Y chromosomes.
- In the creeping vole, Microtus oregoni, the females, with just one X chromosome each, produce X gametes only, and the males, XY, produce Y gametes, or gametes devoid of any sex chromosome, through nondisjunction.
Outside of the rodents, the black muntjac, Muntiacus crinifrons, evolved new X and Y chromosomes through fusions of the ancestral sex chromosomes and autosomes.
1:1 sex ratio
Fisher's principle outlines why almost all species using sexual reproduction have a sex ratio of 1:1. W. D. Hamilton gave the following basic explanation in his 1967 paper on "Extraordinary sex ratios", given the condition that males and females cost equal amounts to produce:
- Suppose male births are less common than female.
- A newborn male then has better mating prospects than a newborn female, and therefore can expect to have more offspring.
- Therefore, parents genetically disposed to produce males tend to have more than average numbers of grandchildren born to them.
- Therefore, the genes for male-producing tendencies spread, and male births become more common.
- As the 1:1 sex ratio is approached, the advantage associated with producing males dies away.
- The same reasoning holds if females are substituted for males throughout. Therefore, 1:1 is the equilibrium ratio.
Non-therian Y chromosome
Many
groups of organisms in addition to therian mammals have Y chromosomes,
but these Y chromosomes do not share common ancestry with therian Y
chromosomes. Such groups include monotremes, Drosophila, some other insects, some fish, some reptiles, and some plants. In Drosophila melanogaster, the Y chromosome does not trigger male development. Instead, sex is determined by the number of X chromosomes. The D. melanogaster Y chromosome does contain genes necessary for male fertility. So XXY D. melanogaster are female, and D. melanogaster with a single X (X0), are male but sterile. There are some species of Drosophila in which X0 males are both viable and fertile.
ZW chromosomes
Other
organisms have mirror image sex chromosomes: where the homogeneous sex
is the male, said to have two Z chromosomes, and the female is the
heterogeneous sex, and said to have a Z chromosome and a W chromosome. For example, female birds, snakes, and butterflies have ZW sex chromosomes, and males have ZZ sex chromosomes.
Non-inverted Y chromosome
There are some species, such as the Japanese rice fish,
the XY system is still developing and cross over between the X and Y is
still possible. Because the male specific region is very small and
contains no essential genes, it is even possible to artificially induce
XX males and YY females to no ill effect.
Multiple XY pairs
Monotremes possess four or five (platypus)
pairs of XY sex chromosomes, each pair consisting of sex chromosomes
with homologous regions. The chromosomes of neighboring pairs are
partially homologous, such that a chain is formed during mitosis.
The first X chromosome in the chain is also partially homologous with
the last Y chromosome, indicating that profound rearrangements, some
adding new pieces from autosomes, have occurred in history.
Platypus sex chromosomes have strong sequence similarity with the avian Z chromosome, (indicating close homology),
and the SRY gene so central to sex-determination in most other mammals
is apparently not involved in platypus sex-determination.
Human Y chromosome
In humans, the Y chromosome spans about 58 million base pairs (the building blocks of DNA) and represents approximately 1% of the total DNA in a male cell. The human Y chromosome contains over 200 genes, at least 72 of which code for proteins. Traits that are inherited via the Y chromosome are called Y-linked, or holandric traits.
Men can lose the Y chromosome in a subset of cells, which is called the mosaic loss of chromosome Y (LOY). This post-zygotic
mutation is strongly associated with age, affecting about 15% of men 70
years of age. Smoking is another important risk factor for LOY. It has been found that men with a higher percentage of hematopoietic stem cells in blood lacking the Y chromosome (and perhaps a higher percentage of other cells lacking it) have a higher risk of certain cancers
and have a shorter life expectancy. Men with LOY (which was defined as
no Y in at least 18% of their hematopoietic cells) have been found to
die 5.5 years earlier on average than others. This has been interpreted
as a sign that the Y chromosome plays a role going beyond sex
determination and reproduction
(although the loss of Y may be an effect rather than a cause). Male
smokers have between 1.5 and 2 times the risk of non-respiratory cancers
as female smokers.
Non-combining region of Y (NRY)
The human Y chromosome is normally unable to recombine with the X chromosome, except for small pieces of pseudoautosomal regions at the telomeres (which comprise about 5% of the chromosome's length). These regions are relics of ancient homology
between the X and Y chromosomes. The bulk of the Y chromosome, which
does not recombine, is called the "NRY", or non-recombining region of
the Y chromosome. The single-nucleotide polymorphisms (SNPs) in this region are used to trace direct paternal ancestral lines.
Genes
Number of genes
The following are some of the gene count estimates of human Y chromosome. Because researchers use different approaches to genome annotation their predictions of the number of genes on each chromosome varies (for technical details, see gene prediction). Among various projects, the collaborative consensus coding sequence project (CCDS)
takes an extremely conservative strategy. So CCDS's gene number
prediction represents a lower bound on the total number of human
protein-coding genes.
| Estimated by | Protein-coding genes | Non-coding RNA genes | Pseudogenes |
|---|---|---|---|
| CCDS | 63 | — | — |
| HGNC | 45 | 55 | 381 |
| Ensembl | 63 | 109 | 392 |
| UniProt | 47 | — | — |
| NCBI | 73 | 122 | 400 |
Gene list
In general, the human Y chromosome is extremely gene poor—it is one of the largest gene deserts in the human genome. Disregarding pseudoautosomal genes, genes encoded on the human Y chromosome include:
- NRY, with corresponding gene on X chromosome
- NRY, other
- AZF1 (azoospermia factor 1)
- BPY2 (basic protein on the Y chromosome)
- DAZ1 (deleted in azoospermia)
- DAZ2
- DFNY1 encoding protein Deafness, Y-linked 1
- PRKY (protein kinase, Y-linked)
- RBMY1A1
- SRY (sex-determining region)
- TSPY (testis-specific protein)
- USP9Y
- UTY (ubiquitously transcribed TPR gene on Y chromosome)
- ZFY (zinc finger protein)
Y-chromosome-linked diseases
Diseases linked to the Y chromosome typically involve an aneuploidy, an atypical number of chromosomes.
Y chromosome microdeletion
Y chromosome microdeletion
(YCM) is a family of genetic disorders caused by missing genes in the Y
chromosome. Many affected men exhibit no symptoms and lead normal
lives. However, YCM is also known to be present in a significant number
of men with reduced fertility or reduced sperm count.
Defective Y chromosome
This results in the person presenting a female phenotype (i.e., is born with female-like genitalia) even though that person possesses an XY karyotype. The lack of the second X results in infertility. In other words, viewed from the opposite direction, the person goes through defeminization but fails to complete masculinization.
The cause can be seen as an incomplete Y chromosome: the usual
karyotype in these cases is 45X, plus a fragment of Y. This usually
results in defective testicular development, such that the infant may or
may not have fully formed male genitalia internally or externally. The
full range of ambiguity of structure may occur, especially if mosaicism is present. When the Y fragment is minimal and nonfunctional, the child is usually a girl with the features of Turner syndrome or mixed gonadal dysgenesis.
XXY
Klinefelter syndrome (47, XXY) is not an aneuploidy
of the Y chromosome, but a condition of having an extra X chromosome,
which usually results in defective postnatal testicular function. The
mechanism is not fully understood; it does not seem to be due to direct
interference by the extra X with expression of Y genes.
XYY
47, XYY syndrome (simply known as XYY syndrome) is caused by the
presence of a single extra copy of the Y chromosome in each of a male's
cells. 47, XYY males have one X chromosome and two Y chromosomes, for a
total of 47 chromosomes per cell. Researchers have found that an extra
copy of the Y chromosome is associated with increased stature and an
increased incidence of learning problems in some boys and men, but the
effects are variable, often minimal, and the vast majority do not know
their karyotype.
In 1965 and 1966 Patricia Jacobs and colleagues published a chromosome survey of 315 male patients at
Scotland's only special security hospital for the developmentally disabled,
finding a higher than expected number of patients to have an extra Y chromosome.
The authors of this study wondered "whether an extra Y chromosome
predisposes its carriers to unusually aggressive behaviour", and this
conjecture "framed the next fifteen years of research on the human Y
chromosome".
Through studies over the next decade, this conjecture was shown
to be incorrect: the elevated crime rate of XYY males is due to lower
median intelligence and not increased aggression, and increased height was the only characteristic that could be reliably associated with XYY males. The "criminal karyotype" concept is therefore inaccurate.
Rare
The following Y-chromosome-linked diseases are rare, but notable because of their elucidating of the nature of the Y chromosome.
More than two Y chromosomes
Greater
degrees of Y chromosome polysomy (having more than one extra copy of
the Y chromosome in every cell, e.g., XYYY) are rare. The extra genetic
material in these cases can lead to skeletal abnormalities, decreased
IQ, and delayed development, but the severity features of these
conditions are variable.
XX male syndrome
XX male syndrome occurs when there has been a recombination in the formation of the male gametes, causing the SRY
portion of the Y chromosome to move to the X chromosome. When such an X
chromosome contributes to the child, the development will lead to a
male, because of the SRY gene.
Genetic genealogy
In human genetic genealogy (the application of genetics to traditional genealogy),
use of the information contained in the Y chromosome is of particular
interest because, unlike other chromosomes, the Y chromosome is passed
exclusively from father to son, on the patrilineal line. Mitochondrial DNA, maternally inherited to both sons and daughters, is used in an analogous way to trace the matrilineal line.
Brain function
Research
is currently investigating whether male-pattern neural development is a
direct consequence of Y-chromosome-related gene expression or an
indirect result of Y-chromosome-related androgenic hormone production.
Microchimerism
The presence of male chromosomes in fetal cells in the blood circulation of women was discovered in 1974.
In 1996, it was found that male fetal progenitor cells could
persist postpartum in the maternal blood stream for as long as 27 years.
A 2004 study at the Fred Hutchinson Cancer Research Center,
Seattle, investigated the origin of male chromosomes found in the
peripheral blood of women who had not had male progeny. A total of 120
subjects (women who had never had sons) were investigated, and it was
found that 21% of them had male DNA. The subjects were categorised into
four groups based on their case histories:
- Group A (8%) had had only female progeny.
- Patients in Group B (22%) had a history of one or more miscarriages.
- Patients Group C (57%) had their pregnancies medically terminated.
- Group D (10%) had never been pregnant before.
The study noted that 10% of the women had never been pregnant before,
raising the question of where the Y chromosomes in their blood could
have come from. The study suggests that possible reasons for occurrence
of male chromosome microchimerism could be one of the following:
- miscarriages,
- pregnancies,
- vanished male twin,
- possibly from sexual intercourse.
A 2012 study at the same institute has detected cells with the Y chromosome in multiple areas of the brains of deceased