From Wikipedia, the free encyclopedia
https://en.wikipedia.org/wiki/Translation_(biology)
In molecular biology and genetics, translation is the process in which ribosomes in the cytoplasm or endoplasmic reticulum synthesize proteins after the process of transcription of DNA to RNA in the cell's nucleus. The entire process is called gene expression.
In translation, messenger RNA (mRNA) is decoded in a ribosome, outside the nucleus, to produce a specific amino acid chain, or polypeptide. The polypeptide later folds into an active protein and performs its functions in the cell. The ribosome facilitates decoding by inducing the binding of complementary tRNA anticodon sequences to mRNA codons. The tRNAs carry specific amino acids that are chained together into a polypeptide as the mRNA passes through and is "read" by the ribosome.
Translation proceeds in three phases:
- Initiation: The ribosome assembles around the target mRNA. The first tRNA is attached at the start codon.
- Elongation: The last tRNA validated by the small ribosomal subunit (accommodation) transfers the amino acid. It carries to the large ribosomal subunit which binds it to the one of the preceding admitted tRNA (transpeptidation). The ribosome then moves to the next mRNA codon to continue the process (translocation), creating an amino acid chain.
- Termination: When a stop codon is reached, the ribosome releases the polypeptide. The ribosomal complex remains intact and moves on to the next mRNA to be translated.
In prokaryotes (bacteria and archaea), translation occurs in the cytosol, where the large and small subunits of the ribosome bind to the mRNA. In eukaryotes, translation occurs in the cytoplasm or across the membrane of the endoplasmic reticulum in a process called co-translational translocation. In co-translational translocation, the entire ribosome/mRNA complex binds to the outer membrane of the rough endoplasmic reticulum (ER), and the new protein is synthesized and released into the ER; the newly created polypeptide can be stored inside the ER for future vesicle transport and secretion outside the cell, or immediately secreted.
Many types of transcribed RNA, such as transfer RNA, ribosomal RNA, and small nuclear RNA, do not undergo a translation into proteins.
Several antibiotics act by inhibiting translation. These include anisomycin, cycloheximide, chloramphenicol, tetracycline, streptomycin, erythromycin, and puromycin. Prokaryotic ribosomes have a different structure from that of eukaryotic ribosomes, and thus antibiotics can specifically target bacterial infections without any harm to a eukaryotic host's cells.
Basic mechanisms
The basic process of protein production is the addition of one amino acid at a time to the end of a protein. This operation is performed by a ribosome. A ribosome is made up of two subunits, a small subunit, and a large subunit. These subunits come together before the translation of mRNA into a protein to provide a location for translation to be carried out and a polypeptide to be produced. The choice of amino acid type to add is determined by an mRNA molecule. Each amino acid added is matched to a three-nucleotide subsequence of the mRNA. For each such triplet possible, the corresponding amino acid is accepted. The successive amino acids added to the chain are matched to successive nucleotide triplets in the mRNA. In this way, the sequence of nucleotides in the template mRNA chain determines the sequence of amino acids in the generated amino acid chain. The addition of an amino acid occurs at the C-terminus of the peptide; thus, translation is said to be amine-to-carboxyl directed.
The mRNA carries genetic information encoded as a ribonucleotide sequence from the chromosomes to the ribosomes. The ribonucleotides are "read" by translational machinery in a sequence of nucleotide triplets called codons. Each of those triplets codes for a specific amino acid.
The ribosome molecules translate this code to a specific sequence of amino acids. The ribosome is a multisubunit structure containing rRNA and proteins. It is the "factory" where amino acids are assembled into proteins. tRNAs are small noncoding RNA chains (74–93 nucleotides) that transport amino acids to the ribosome. The repertoire of tRNA genes varies widely between species, with some Bacteria having between 20 and 30 genes while complex eukaryotes could have thousands. tRNAs have a site for amino acid attachment, and a site called an anticodon. The anticodon is an RNA triplet complementary to the mRNA triplet that codes for their cargo amino acid.
Aminoacyl tRNA synthetases (enzymes) catalyze the bonding between specific tRNAs and the amino acids that their anticodon sequences call for. The product of this reaction is an aminoacyl-tRNA. In bacteria, this aminoacyl-tRNA is carried to the ribosome by EF-Tu, where mRNA codons are matched through complementary base pairing to specific tRNA anticodons. Aminoacyl-tRNA synthetases that mispair tRNAs with the wrong amino acids can produce mischarged aminoacyl-tRNAs, which can result in inappropriate amino acids at the respective position in the protein. This "mistranslation" of the genetic code naturally occurs at low levels in most organisms, but certain cellular environments cause an increase in permissive mRNA decoding, sometimes to the benefit of the cell.
The ribosome has two binding sites for tRNA. They are the aminoacyl site (abbreviated A), and the peptidyl site/ exit site (abbreviated P/E). Concerning the mRNA, the three sites are oriented 5’ to 3’ E-P-A, because ribosomes move toward the 3' end of mRNA. The A-site binds the incoming tRNA with the complementary codon on the mRNA. The P/E-site holds the tRNA with the growing polypeptide chain. When an aminoacyl-tRNA initially binds to its corresponding codon on the mRNA, it is in the A site. Then, a peptide bond forms between the amino acid of the tRNA in the A site and the amino acid of the charged tRNA in the P/E site. The growing polypeptide chain is transferred to the tRNA in the A site. Translocation occurs, moving the tRNA to the P/E site, now without an amino acid; the tRNA that was in the A site, now charged with the polypeptide chain, is moved to the P/E site and the tRNA leaves, and nothinger aminoacyl-tRNA enters the A site to repeat the process.
After the new amino acid is added to the chain, and after the tRNA is released out of the ribosome and into the cytosol, the energy provided by the hydrolysis of a GTP bound to the translocase EF-G (in bacteria) and a/eEF-2 (in eukaryotes and archaea) moves the ribosome down one codon towards the 3' end. The energy required for translation of proteins is significant. For a protein containing n amino acids, the number of high-energy phosphate bonds required to translate it is 4n-1. The rate of translation varies; it is significantly higher in prokaryotic cells (up to 17–21 amino acid residues per second) than in eukaryotic cells (up to 6–9 amino acid residues per second).
Even though the ribosomes are usually considered accurate and processive machines, the translation process is subject to errors that can lead either to the synthesis of erroneous proteins or to the premature abandonment of translation, either because a tRNA couples to a wrong codon or because a tRNA is coupled to the wrong amino acid. The rate of error in synthesizing proteins has been estimated to be between 1 in 105 and 1 in 103 misincorporated amino acids, depending on the experimental conditions. The rate of premature translation abandonment, instead, has been estimated to be of the order of magnitude of 10−4 events per translated codon. The correct amino acid is covalently bonded to the correct transfer RNA (tRNA) by amino acyl transferases. The amino acid is joined by its carboxyl group to the 3' OH of the tRNA by an ester bond. When the tRNA has an amino acid linked to it, the tRNA is termed "charged". Initiation involves the small subunit of the ribosome binding to the 5' end of mRNA with the help of initiation factors (IF). In bacteria and a minority of archaea, initiation of protein synthesis involves the recognition of a purine-rich initiation sequence on the mRNA called the Shine-Dalgarno sequence. The Shine-Dalgarno sequence binds to a complementary pyrimidine-rich sequence on the 3' end of the 16S rRNA part of the 30S ribosomal subunit. The binding of these complementary sequences ensures that the 30S ribosomal subunit is bound to the mRNA and is aligned such that the initiation codon is placed in the 30S portion of the P-site. Once the mRNA and 30S subunit are properly bound, an initiation factor brings the initiator tRNA-amino acid complex, f-Met-tRNA, to the 30S P site. The initiation phase is completed once a 50S subunit joins the 30 subunit, forming an active 70S ribosome. Termination of the polypeptide occurs when the A site of the ribosome is occupied by a stop codon (UAA, UAG, or UGA) on the mRNA, creating the primary structure of a protein. tRNA usually cannot recognize or bind to stop codons. Instead, the stop codon induces the binding of a release factor protein (RF1 & RF2) that prompts the disassembly of the entire ribosome/mRNA complex by the hydrolysis of the polypeptide chain from the peptidyl transferase center of the ribosome. Drugs or special sequence motifs on the mRNA can change the ribosomal structure so that near-cognate tRNAs are bound to the stop codon instead of the release factors. In such cases of 'translational readthrough', translation continues until the ribosome encounters the next stop codon.
The process of translation is highly regulated in both eukaryotic and prokaryotic organisms. Regulation of translation can impact the global rate of protein synthesis which is closely coupled to the metabolic and proliferative state of a cell. In addition, recent work has revealed that genetic differences and their subsequent expression as mRNAs can also impact translation rate in an RNA-specific manner.
Clinical significance
Translational control is critical for the development and survival of cancer. Cancer cells must frequently regulate the translation phase of gene expression, though it is not fully understood why translation is targeted over steps like transcription. While cancer cells often have genetically altered translation factors, it is much more common for cancer cells to modify the levels of existing translation factors. Several major oncogenic signaling pathways, including the RAS–MAPK, PI3K/AKT/mTOR, MYC, and WNT–β-catenin pathways, ultimately reprogram the genome via translation. Cancer cells also control translation to adapt to cellular stress. During stress, the cell translates mRNAs that can mitigate the stress and promote survival. An example of this is the expression of AMPK in various cancers; its activation triggers a cascade that can ultimately allow the cancer to escape apoptosis (programmed cell death) triggered by nutrition deprivation. Future cancer therapies may involve disrupting the translation machinery of the cell to counter the downstream effects of cancer.
Mathematical modeling of translation
The transcription-translation process description, mentioning only the most basic ”elementary” processes, consists of:
- production of mRNA molecules (including splicing),
- initiation of these molecules with help of initiation factors (e.g., the initiation can include the circularization step though it is not universally required),
- initiation of translation, recruiting the small ribosomal subunit,
- assembly of full ribosomes,
- elongation, (i.e. movement of ribosomes along mRNA with production of protein),
- termination of translation,
- degradation of mRNA molecules,
- degradation of proteins.
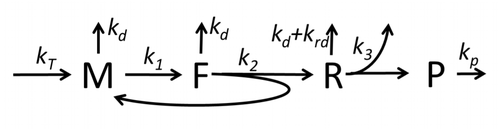
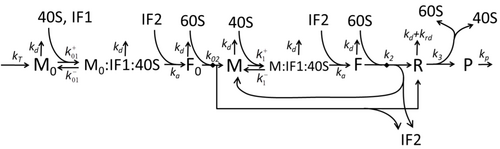
The process of amino acid building to create protein in translation is a subject of various physic models for a long time starting from the first detailed kinetic models such as or others taking into account stochastic aspects of translation and using computer simulations. Many chemical kinetics-based models of protein synthesis have been developed and analyzed in the last four decades. Beyond chemical kinetics, various modeling formalisms such as Totally Asymmetric Simple Exclusion Process (TASEP),[23]Probabilistic Boolean Networks (PBN), Petri Nets and max-plus algebra have been applied to model the detailed kinetics of protein synthesis or some of its stages. A basic model of protein synthesis that takes into account all eight 'elementary' processes has been developed, following the paradigm that "useful models are simple and extendable". The simplest model M0 is represented by the reaction kinetic mechanism (Figure M0). It was generalised to include 40S, 60S and initiation factors (IF) binding (Figure M1'). It was extended further to include effect of microRNA on protein synthesis. Most of models in this hierarchy can be solved analytically. These solutions were used to extract 'kinetic signatures' of different specific mechanisms of synthesis regulation.
Genetic code
It is also possible to translate either by hand (for short sequences) or by computer (after first programming one appropriately, see section below); this allows biologists and chemists to draw out the chemical structure of the encoded protein on paper.
First, convert each template DNA base to its RNA complement (note that the complement of A is now U), as shown below. Note that the template strand of the DNA is the one the RNA is polymerized against; the other DNA strand would be the same as the RNA, but with thymine instead of uracil.
DNA -> RNA A -> U T -> A C -> G G -> C A=T-> A=U
Then split the RNA into triplets (groups of three bases). Note that there are 3 translation "windows", or reading frames, depending on where you start reading the code. Finally, use the table at Genetic code to translate the above into a structural formula as used in chemistry.
This will give you the primary structure of the protein. However, proteins tend to fold, depending in part on hydrophilic and hydrophobic segments along the chain. Secondary structure can often still be guessed at, but the proper tertiary structure is often very hard to determine.
Whereas other aspects such as the 3D structure, called tertiary structure, of protein can only be predicted using sophisticated algorithms, the amino acid sequence, called primary structure, can be determined solely from the nucleic acid sequence with the aid of a translation table.
This approach may not give the correct amino acid composition of the protein, in particular if unconventional amino acids such as selenocysteine are incorporated into the protein, which is coded for by a conventional stop codon in combination with a downstream hairpin (SElenoCysteine Insertion Sequence, or SECIS).
There are many computer programs capable of translating a DNA/RNA sequence into a protein sequence. Normally this is performed using the Standard Genetic Code, however, few programs can handle all the "special" cases, such as the use of the alternative initiation codons which are biologically significant. For instance, the rare alternative start codon CTG codes for Methionine when used as a start codon, and for Leucine in all other positions.
Example: Condensed translation table for the Standard Genetic Code (from the NCBI Taxonomy webpage).
AAs = FFLLSSSSYY**CC*WLLLLPPPPHHQQRRRRIIIMTTTTNNKKSSRRVVVVAAAADDEEGGGG Starts = ---M---------------M---------------M---------------------------- Base1 = TTTTTTTTTTTTTTTTCCCCCCCCCCCCCCCCAAAAAAAAAAAAAAAAGGGGGGGGGGGGGGGG Base2 = TTTTCCCCAAAAGGGGTTTTCCCCAAAAGGGGTTTTCCCCAAAAGGGGTTTTCCCCAAAAGGGG Base3 = TCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAG
The "Starts" row indicate three start codons, UUG, CUG, and the very common AUG. It also indicates the first amino acid residue when interpreted as a start: in this case it is all methionine.
Translation tables
Even when working with ordinary eukaryotic sequences such as the Yeast genome, it is often desired to be able to use alternative translation tables—namely for translation of the mitochondrial genes. Currently the following translation tables are defined by the NCBI Taxonomy Group for the translation of the sequences in GenBank:
- The standard code
- The vertebrate mitochondrial code
- The yeast mitochondrial code
- The mold, protozoan, and coelenterate mitochondrial code and the mycoplasma/spiroplasma code
- The invertebrate mitochondrial code
- The ciliate, dasycladacean and hexamita nuclear code
- The kinetoplast code
- The echinoderm and flatworm mitochondrial code
- The euplotid nuclear code
- The bacterial, archaeal and plant plastid code
- The alternative yeast nuclear code
- The ascidian mitochondrial code
- The alternative flatworm mitochondrial code
- The Blepharisma nuclear code
- The chlorophycean mitochondrial code
- The trematode mitochondrial code
- The Scenedesmus obliquus mitochondrial code
- The Thraustochytrium mitochondrial code
- The Pterobranchia mitochondrial code
- The candidate division SR1 and gracilibacteria code
- The Pachysolen tannophilus nuclear code
- The karyorelict nuclear code
- The Condylostoma nuclear code
- The Mesodinium nuclear code
- The peritrich nuclear code
- The Blastocrithidia nuclear code
- The Cephalodiscidae mitochondrial code



















![{\displaystyle \rho _{X,Y}=\operatorname {corr} (X,Y)={\operatorname {cov} (X,Y) \over \sigma _{X}\sigma _{Y}}={\operatorname {E} [(X-\mu _{X})(Y-\mu _{Y})] \over \sigma _{X}\sigma _{Y}},\quad {\text{if}}\ \sigma _{X}\sigma _{Y}>0.}](https://wikimedia.org/api/rest_v1/media/math/render/svg/b551ad29592ae746bf05fe397fbdc56201f483a5)
















![{\displaystyle {\begin{aligned}r_{xy}&={\frac {\sum x_{i}y_{i}-n{\bar {x}}{\bar {y}}}{ns'_{x}s'_{y}}}\\[5pt]&={\frac {n\sum x_{i}y_{i}-\sum x_{i}\sum y_{i}}{{\sqrt {n\sum x_{i}^{2}-(\sum x_{i})^{2}}}~{\sqrt {n\sum y_{i}^{2}-(\sum y_{i})^{2}}}}}.\end{aligned}}}](https://wikimedia.org/api/rest_v1/media/math/render/svg/6da33b8144a5e67959969ef2c4830ece1938bbb2)











![{\displaystyle {\begin{aligned}\rho _{X,Y}&={\frac {1}{\sigma _{X}\sigma _{Y}}}\mathrm {E} [(X-\mu _{X})(Y-\mu _{Y})]\\[5pt]&={\frac {1}{\sigma _{X}\sigma _{Y}}}\sum _{x,y}{(x-\mu _{X})(y-\mu _{Y})\mathrm {P} (X=x,Y=y)}\\[5pt]&=\left(1-{\frac {2}{3}}\right)(-1-0){\frac {1}{3}}+\left(0-{\frac {2}{3}}\right)(0-0){\frac {1}{3}}+\left(1-{\frac {2}{3}}\right)(1-0){\frac {1}{3}}=0.\end{aligned}}}](https://wikimedia.org/api/rest_v1/media/math/render/svg/75bf2b7806338758b4c55d7b4f18a5071b8e919b)

![{\displaystyle [0,+\infty ]}](https://wikimedia.org/api/rest_v1/media/math/render/svg/f32245981f739c86ea8f68ce89b1ad6807428d35)
![[-1, 1]](https://wikimedia.org/api/rest_v1/media/math/render/svg/51e3b7f14a6f70e614728c583409a0b9a8b9de01)