In human genetics, the Y-chromosomal most recent common ancestor (Y-MRCA, informally known as Y-chromosomal Adam) is the most recent common ancestor (MRCA) from whom all currently living men are descended patrilineally. The term Y-MRCA reflects the fact that the Y chromosomes of all currently living human males are directly derived from the Y chromosome of this remote ancestor. The analogous concept of the matrilineal most recent common ancestor is known as "Mitochondrial Eve" (mt-MRCA, named for the matrilineal transmission of mtDNA), the most recent woman from whom all living humans are descended matrilineally. As with "Mitochondrial Eve", the title of "Y-chromosomal Adam" is not permanently fixed to a single individual, but can advance over the course of human history as paternal lineages become extinct.
Estimates of the time when Y-MRCA lived have also shifted as modern knowledge of human ancestry changes. In 2013, the discovery of a previously unknown Y-chromosomal haplogroup was announced, which resulted in a slight adjustment of the estimated age of the human Y-MRCA.
By definition, it is not necessary that the Y-MRCA and the mt-MRCA should have lived at the same time. While estimates as of 2014 suggested the possibility that the two individuals may well have been roughly contemporaneous (albeit with uncertainties ranging in the tens of thousands of years), the discovery of archaic Y-haplogroup has pushed back the estimated age of the Y-MRCA beyond the most likely age of the mt-MRCA. As of 2015, estimates of the age of the Y-MRCA range around 200,000 to 300,000 years ago, roughly consistent with the emergence of anatomically modern humans.
Y-chromosomal data taken from a Neanderthal from El Sidrón, Spain, produced a Y-T-MRCA of 588,000 years ago for neanderthal and Homo sapiens patrilineages, dubbed ante Adam and 275,000 years ago for Y-MRCA.
Definition
The Y-chromosomal most recent common ancestor is the most recent common ancestor of the Y-chromosomes found in currently living human males.
Due to the definition via the "currently living" population, the
identity of a MRCA, and by extension of the human Y-MRCA, is
time-dependent (it depends on the moment in time intended by the term
"currently").
The MRCA of a population may move forward in time as archaic lineages
within the population go extinct:
once a lineage has died out, it is irretrievably lost. This mechanism
can thus only shift the title of Y-MRCA forward in time. Such an event
could be due to the total extinction of several basal haplogroups.
The same holds for the concepts of matrilineal and patrilineal MRCAs: it
follows from the definition of Y-MRCA that he had at least two sons who
both have unbroken lineages that have survived to the present day. If
the lineages of all but one of those sons die out, then the title of
Y-MRCA shifts forward from the remaining son through his patrilineal
descendants, until the first descendant is reached who had at least two
sons who both have living, patrilineal descendants. The title of Y-MRCA
is not permanently fixed to a single individual, and the Y-MRCA for any
given population would himself have been part of a population which had
its own, more remote, Y-MRCA.
Although the informal name "Y-chromosomal Adam" is a reference to the biblical Adam, this should not be misconstrued as implying that the bearer of the chromosome was the only human male alive during his time.
His other male contemporaries may also have descendants alive today, but
not, by definition, through solely patrilineal descent; in other words,
none of them have an unbroken male line of descendants (son's son's son's … son) connecting them to currently living people.
By the nature of the concept of most recent common ancestors, these estimates can only represent a terminus ante quem
("limit before which"), until the genome of the entire population has
been examined (in this case, the genome of all living humans).
Age estimate
Estimates
on the age of the Y-MRCA crucially depend on the most archaic known
haplogroup extant in contemporary populations. As of 2018, this is haplogroup A00
(discovered in 2013). Age estimates based on this published during
2014–2015 range between 160,000 and 300,000 years, compatible with the
time of emergence and early dispersal of Homo sapiens.
Method
In
addition to the tendency of the title of Y-MRCA to shift forward in
time, the estimate of the Y-MRCA's DNA sequence, his position in the
family tree, the time when he lived, and his place of origin, are all
subject to future revisions.
The following events would change the estimate of who the individual designated as Y-MRCA was:
- Further sampling of Y chromosomes could uncover previously unknown divergent lineages. If this happens, Y-chromosome lineages would converge on an individual who lived further back in time.
- The discovery of additional deep rooting mutations in known lineages could lead to a rearrangement of the family tree.
- Revision of the Y-chromosome mutation rate (see below) can change the estimate of the time when he lived.
The time when Y-MRCA lived is determined by applying a molecular clock to human Y-chromosomes. In contrast to mitochondrial DNA (mtDNA), which has a short sequence of 16,000 base pairs,
and mutates frequently, the Y chromosome is significantly longer at 60
million base pairs, and has a lower mutation rate. These features of the
Y chromosome have slowed down the identification of its polymorphisms;
as a consequence, they have reduced the accuracy of Y-chromosome
mutation rate estimates.
Methods of estimating the age of the Y-MRCA for a population of
human males whose Y-chromosomes have been sequenced are based on
applying the theories of molecular evolution to the Y chromosome. Unlike the autosomes, the human Y-chromosome does not recombine often with the X chromosome during meiosis,
but is usually transferred intact from father to son; however, it can
recombine with the X chromosome in the pseudoautosomal regions at the
ends of the Y chromosome. Mutations occur periodically within the Y chromosome, and these mutations are passed on to males in subsequent generations.
These mutations can be used as markers to identify shared
patrilineal relationships. Y chromosomes that share a specific mutation
are referred to as haplogroups.
Y chromosomes within a specific haplogroup are assumed to share a
common patrilineal ancestor who was the first to carry the defining
mutation. (This assumption could be mistaken, as it is possible for the
same mutation to occur more than once.) A family tree of Y chromosomes
can be constructed, with the mutations serving as branching points along
lineages. The Y-MRCA is positioned at the root of the family tree, as
the Y chromosomes of all living males are descended from his Y
chromosome.
Researchers can reconstruct ancestral Y chromosome DNA sequences
by reversing mutated DNA segments to their original condition. The most
likely original or ancestral state of a DNA sequence is determined by
comparing human DNA sequences with those of a closely related species,
usually non-human primates such as chimpanzees and gorillas. By
reversing known mutations in a Y-chromosome lineage, a hypothetical
ancestral sequence for the MRCA, Y-chromosomal Adam, can be inferred.
Determining the Y-MRCA's DNA sequence, and the time when he
lived, involves identifying the human Y-chromosome lineages that are
most divergent from each other—the lineages that share the fewest
mutations with each other when compared to a non-human primate sequence
in a phylogenetic tree. The common ancestor of the most divergent lineages is therefore the common ancestor of all lineages.
History of estimates
Early estimates of the age for the Y-MRCA published during the 1990s ranged between roughly 200 and 300 kya,
Such estimates were later substantially revised downward, as in Thomson et al. 2000, which proposed an age of about 59,000.
This date suggested that the Y-MRCA lived about 84,000 years after his female counterpart mt-MRCA (the matrilineal most recent common ancestor), who lived 150,000–200,000 years ago.
This date also meant that Y-chromosomal Adam lived at a time very close to, and possibly after, the migration from Africa
which is believed to have taken place 50,000–80,000 years ago.
One explanation given for this discrepancy in the time depths of
patrilineal vs. matrilineal lineages was that females have a better
chance of reproducing than males due to the practice of polygyny.
When a male individual has several wives, he has effectively prevented
other males in the community from reproducing and passing on their Y
chromosomes to subsequent generations. On the other hand, polygyny does
not prevent most females in a community from passing on their
mitochondrial DNA to subsequent generations. This differential
reproductive success of males and females can lead to fewer male
lineages relative to female lineages persisting into the future. These
fewer male lineages are more sensitive to drift and would most likely coalesce on a more recent common ancestor. This would potentially explain the more recent dates associated with the Y-MRCA.
The "hyper-recent" estimate of significantly below 100 kya was
again corrected upward in studies of the early 2010s, which ranged at
about 120 kya to 160 kya.
This revision was due to the discovery of additional mutations and the
rearrangement of the backbone of the Y-chromosome phylogeny following
the resequencing of Haplogroup A lineages.
In 2013, Francalacci et al. reported the sequencing of male-specific single-nucleotide Y-chromosome polymorphisms (MSY-SNPs) from 1204 Sardinian men, which indicated an estimate of 180,000 to 200,000 years for the common origin of all humans through paternal lineage. or again as high as 180 to 200 kya.
Also in 2013, Poznik et al. reported the Y-MRCA to have lived between 120,000 and 156,000 years ago, based on genome sequencing of 69 men from 9 different populations.
In addition, the same study estimated the age of Mitochondrial Eve to about 99,000 and 148,000 years.
As these ranges overlap for a time-range of 28,000 years (148 to 120
kya), the results of this study have been cast in terms of the
possibility that "Genetic Adam and Eve may have walked on Earth at the
same time" in the popular press.
The announcement of yet another discovery of a previously unknown lineage, haplogroup A00,
in 2013, resulted in another shift in the estimate for the age of
Y-chromosomal. Elhaik et al. (2014) dated it to between 163,900 and
260,200 years ago (95% CI). Karmin et al. (2015) dated it to between 192,000 and 307,000 years ago (95% CI). The same study reports that non-African populations converge to a cluster of Y-MRCAs in a window close to 50kya (out-of-Africa migration), and an additional bottleneck
for non-African populations at about 10kya, interpreted as reflecting
cultural changes increasing the variance in male reproductive success
(i.e. increased social stratification) in the Neolithic.
Family tree
The
revised root of the y-chromosome family tree by Cruciani et al. 2011
compared with the family tree from Karafet et al. 2008. This has been
further expanded by the discoveries published by Mendez et al. in 2013.
Initial sequencing (Karafet et al., 2008) of the human Y chromosome suggested that two most basal Y-chromosome lineages were Haplogroup A and Haplogroup BT.
Haplogroup A is found at low frequencies in parts of Africa, but is
common among certain hunter-gatherer groups. Haplogroup BT lineages
represent the majority of African Y-chromosome lineages and virtually
all non-African lineages.
Y-chromosomal Adam was represented as the root of these two lineages.
Haplogroup A and Haplogroup BT represented the lineages of the two male
descendants of Y-chromosomal Adam.
Cruciani et al. 2011, determined that the deepest split in the
Y-chromosome tree was found between two previously reported subclades of
Haplogroup A, rather than between Haplogroup A and Haplogroup BT.
Subclades A1b and A1a-T are now believed to descend directly from the
root of the tree and now represent the lineages of Y-chromosomal Adam's
two sons. The rearrangement of the Y-chromosome family tree implies that
lineages classified as Haplogroup A do not necessarily form a monophyletic clade.
Haplogroup A therefore refers to a collection of lineages that do not
possess the markers that define Haplogroup BT, though Haplogroup A
includes the most distantly related Y chromosomes.
The M91 and P97 mutations distinguish Haplogroup A from
Haplogroup BT. Within Haplogroup A chromosomes, the M91 marker consists
of a stretch of 8 T nucleobase units. In Haplogroup BT and chimpanzee
chromosomes, this marker consists of 9 T nucleobase units. This pattern
suggested that the 9T stretch of Haplogroup BT was the ancestral
version and that Haplogroup A was formed by the deletion of one nucleobase. Haplogroups A1b and A1a were considered subclades of Haplogroup A as they both possessed the M91 with 8Ts.
But according to Cruciani et al. 2011, the region surrounding the
M91 marker is a mutational hotspot prone to recurrent mutations. It is
therefore possible that the 8T stretch of Haplogroup A may be the
ancestral state of M91 and the 9T of Haplogroup BT may be the derived
state that arose by an insertion
of 1T. This would explain why subclades A1b and A1a-T, the deepest
branches of Haplogroup A, both possess the same version of M91 with 8Ts.
Furthermore, Cruciani et al. 2011 determined that the P97 marker, which
is also used to identify Haplogroup A, possessed the ancestral state in
Haplogroup A but the derived state in Haplogroup BT.
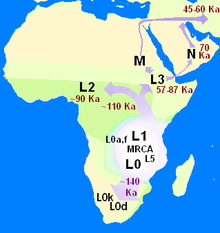
Likely geographic origin
As current estimates on TMRCA converge with estimates for the age of anatomically modern humans
and well predate the Out of Africa migration, geographical origin hypotheses continue to be limited to the African continent.
According to Cruciani et al. 2011, the most basal lineages have been detected in West, Northwest and Central Africa, suggesting plausibility for the Y-MRCA living in the general region of "Central-Northwest Africa".
Scozzari et al. (2012) agreed with a plausible placement
in "the north-western quadrant of the African continent" for the
emergence of the A1b haplogroup. The 2013 report of haplogroup A00 found among the Mbo people of western present-day Cameroon is also compatible with this picture.
The revision of Y-chromosomal phylogeny since 2011 has affected
estimates for the likely geographical origin of Y-MRCA as well as
estimates on time depth. By the same reasoning, future discovery of
presently-unknown archaic haplogroups in living people would again lead
to such revisions. In particular, the possible presence of between 1%
and 4% Neanderthal-derived DNA
in Eurasian genomes implies that the (unlikely) event of a discovery of
a single living Eurasian male exhibiting a Neanderthal patrilineal line
would immediately push back T-MRCA ("time to MRCA") to at least twice
its current estimate. However, the discovery of a neanderthal
Y-chromosome by Mendez et al.
suggests the extinction of neanderthal patrilineages, as the lineage
inferred from the neanderthal sequence is outside of the range of contemporary human genetic variation. Questions of geographical origin would become part of the debate on Neanderthal evolution from Homo erectus.