| Cell | |
|---|---|
 A eukaryotic cell (left) and prokaryotic cell (right) |
The cell is the basic structural and functional unit of all forms of life. Every cell consists of cytoplasm enclosed within a membrane; many cells contain organelles, each with a specific function. The term comes from the Latin word cellula meaning 'small room'. Most cells are only visible under a microscope. Cells emerged on Earth about 4 billion years ago. All cells are capable of replication, protein synthesis, and motility.
Cells are broadly categorized into two types: eukaryotic cells, which possess a nucleus, and prokaryotic cells, which lack a nucleus but have a nucleoid region. Prokaryotes are single-celled organisms such as bacteria, whereas eukaryotes can be either single-celled, such as amoebae, or multicellular, such as some algae, plants, animals, and fungi. Eukaryotic cells contain organelles including mitochondria, which provide energy for cell functions; chloroplasts, which create sugars by photosynthesis, in plants; and ribosomes, which synthesise proteins.
Cells were discovered by Robert Hooke in 1665, who named them after their resemblance to cells inhabited by Christian monks in a monastery. Cell theory, developed in 1839 by Matthias Jakob Schleiden and Theodor Schwann, states that all organisms are composed of one or more cells, that cells are the fundamental unit of structure and function in all living organisms, and that all cells come from pre-existing cells.
Cell types
Cells are broadly categorized into two types: eukaryotic cells, which possess a nucleus, and prokaryotic cells, which lack a nucleus but have a nucleoid region. Prokaryotes are single-celled organisms, whereas eukaryotes can be either single-celled or multicellular.
Prokaryotic cells

Prokaryotes include bacteria and archaea, two of the three domains of life. Prokaryotic cells were the first form of life on Earth, characterized by having vital biological processes including cell signaling. They are simpler and smaller than eukaryotic cells, and lack a nucleus, and other membrane-bound organelles. The DNA of a prokaryotic cell consists of a single circular chromosome that is in direct contact with the cytoplasm. The nuclear region in the cytoplasm is called the nucleoid. Most prokaryotes are the smallest of all organisms, ranging from 0.5 to 2.0 μm in diameter.
A prokaryotic cell has three regions:
- Enclosing the cell is the cell envelope, generally consisting of a plasma membrane covered by a cell wall which, for some bacteria, may be further covered by a third layer called a capsule. Though most prokaryotes have both a cell membrane and a cell wall, there are exceptions such as Mycoplasma (bacteria) and Thermoplasma (archaea) which only possess the cell membrane layer. The envelope gives rigidity to the cell and separates the interior of the cell from its environment, serving as a protective filter. The cell wall consists of peptidoglycan in bacteria and acts as an additional barrier against exterior forces. It also prevents the cell from expanding and bursting (cytolysis) from osmotic pressure due to a hypotonic environment. Some eukaryotic cells (plant cells and fungal cells) also have a cell wall.
- Inside the cell is the cytoplasmic region that contains the genome (DNA), ribosomes and various sorts of inclusions. The genetic material is freely found in the cytoplasm. Prokaryotes can carry extrachromosomal DNA elements called plasmids, which are usually circular. Linear bacterial plasmids have been identified in several species of spirochete bacteria, including members of the genus Borrelia notably Borrelia burgdorferi, which causes Lyme disease. Though not forming a nucleus, the DNA is condensed in a nucleoid. Plasmids encode additional genes, such as antibiotic resistance genes.
- On the outside, some prokaryotes have flagella and pili that project from the cell's surface. These are structures made of proteins that facilitate movement and communication between cells.
Eukaryotic cells


Plants, animals, fungi, slime moulds, protozoa, and algae are all eukaryotic. These cells are about fifteen times wider than a typical prokaryote and can be as much as a thousand times greater in volume. The main distinguishing feature of eukaryotes as compared to prokaryotes is compartmentalization: the presence of membrane-bound organelles (compartments) in which specific activities take place. Most important among these is a cell nucleus, an organelle that houses the cell's DNA. This nucleus gives the eukaryote its name, which means "true kernel (nucleus)". Some of the other differences are:
- The plasma membrane resembles that of prokaryotes in function, with minor differences in the setup. Cell walls may or may not be present.
- The eukaryotic DNA is organized in one or more linear molecules, called chromosomes, which are associated with histone proteins. All chromosomal DNA is stored in the cell nucleus, separated from the cytoplasm by a membrane. Some eukaryotic organelles such as mitochondria also contain some DNA.
- Many eukaryotic cells are ciliated with primary cilia. Primary cilia play important roles in chemosensation, mechanosensation, and thermosensation. Each cilium may thus be "viewed as a sensory cellular antennae that coordinates a large number of cellular signaling pathways, sometimes coupling the signaling to ciliary motility or alternatively to cell division and differentiation."
- Motile eukaryotes can move using motile cilia or flagella. Motile cells are absent in conifers and flowering plants. Eukaryotic flagella are more complex than those of prokaryotes.
|
|
Prokaryotes | Eukaryotes |
|---|---|---|
| Typical organisms | bacteria, archaea | protists, algae, fungi, plants, animals |
| Typical size | ~ 1–5 μm | ~ 10–100 μm |
| Type of nucleus | nucleoid region; no true nucleus | true nucleus with double membrane |
| DNA | circular (usually) | linear molecules (chromosomes) with histone proteins |
| RNA/protein synthesis | coupled in the cytoplasm | RNA synthesis in the nucleus protein synthesis in the cytoplasm |
| Ribosomes | 50S and 30S | 60S and 40S |
| Cytoplasmic structure | very few structures | highly structured by endomembranes and a cytoskeleton |
| Cell movement | flagella made of flagellin | flagella and cilia containing microtubules; lamellipodia and filopodia containing actin |
| Mitochondria | none | one to several thousand |
| Chloroplasts | none | in algae and plants |
| Organization | usually single cells | single cells, colonies, higher multicellular organisms with specialized cells |
| Cell division | binary fission (simple division) | mitosis (fission or budding) meiosis |
| Chromosomes | single chromosome | more than one chromosome |
| Membranes | cell membrane | Cell membrane and membrane-bound organelles |
Many groups of eukaryotes are single-celled. Among the many-celled groups are animals and plants. The number of cells in these groups vary with species; it has been estimated that the human body contains around 37 trillion (3.72×1013) cells, and more recent studies put this number at around 30 trillion (~36 trillion cells in the male, ~28 trillion in the female).
Subcellular components
All cells, whether prokaryotic or eukaryotic, have a membrane that envelops the cell, regulates what moves in and out (selectively permeable), and maintains the electric potential of the cell. Inside the membrane, the cytoplasm takes up most of the cell's volume. Except red blood cells, which lack a cell nucleus and most organelles to accommodate maximum space for hemoglobin, all cells possess DNA, the hereditary material of genes, and RNA, containing the information necessary to build various proteins such as enzymes, the cell's primary machinery. There are also other kinds of biomolecules in cells. This article lists these primary cellular components, then briefly describes their function.
Cell membrane

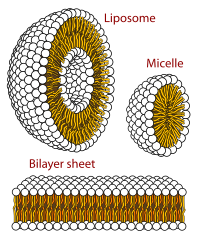
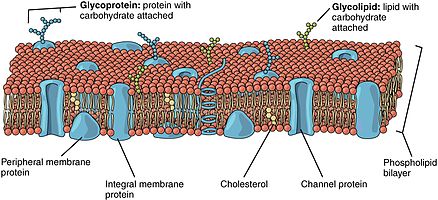
The cell membrane, or plasma membrane, is a selectively permeable biological membrane that surrounds the cytoplasm of a cell. In animals, the plasma membrane is the outer boundary of the cell, while in plants and prokaryotes it is usually covered by a cell wall. This membrane serves to separate and protect a cell from its surrounding environment and is made mostly from a double layer of phospholipids, which are amphiphilic (partly hydrophobic and partly hydrophilic). Hence, the layer is called a phospholipid bilayer, or sometimes a fluid mosaic membrane. Embedded within this membrane is a macromolecular structure called the porosome the universal secretory portal in cells and a variety of protein molecules that act as channels and pumps that move different molecules into and out of the cell. The membrane is semi-permeable, and selectively permeable, in that it can either let a substance (molecule or ion) pass through freely, to a limited extent or not at all. Cell surface membranes also contain receptor proteins that allow cells to detect external signaling molecules such as hormones.
Cytoskeleton

The cytoskeleton acts to organize and maintain the cell's shape; anchors organelles in place; helps during endocytosis, the uptake of external materials by a cell, and cytokinesis, the separation of daughter cells after cell division; and moves parts of the cell in processes of growth and mobility. The eukaryotic cytoskeleton is composed of microtubules, intermediate filaments and microfilaments. In the cytoskeleton of a neuron the intermediate filaments are known as neurofilaments. There are a great number of proteins associated with them, each controlling a cell's structure by directing, bundling, and aligning filaments. The prokaryotic cytoskeleton is less well-studied but is involved in the maintenance of cell shape, polarity and cytokinesis. The subunit protein of microfilaments is a small, monomeric protein called actin. The subunit of microtubules is a dimeric molecule called tubulin. Intermediate filaments are heteropolymers whose subunits vary among the cell types in different tissues. Some of the subunit proteins of intermediate filaments include vimentin, desmin, lamin (lamins A, B and C), keratin (multiple acidic and basic keratins), and neurofilament proteins (NF–L, NF–M).
Genetic material

Two different kinds of genetic material exist: deoxyribonucleic acid (DNA) and ribonucleic acid (RNA). Cells use DNA for their long-term information storage. The biological information contained in an organism is encoded in its DNA sequence. RNA is used for information transport (e.g., mRNA) and enzymatic functions (e.g., ribosomal RNA). Transfer RNA (tRNA) molecules are used to add amino acids during protein translation.
Prokaryotic genetic material is organized in a simple circular bacterial chromosome in the nucleoid region of the cytoplasm. Eukaryotic genetic material is divided into different, linear molecules called chromosomes inside a discrete nucleus, usually with additional genetic material in some organelles like mitochondria and chloroplasts (see endosymbiotic theory).
A human cell has genetic material contained in the cell nucleus (the nuclear genome) and in the mitochondria (the mitochondrial genome). In humans, the nuclear genome is divided into 46 linear DNA molecules called chromosomes, including 22 homologous chromosome pairs and a pair of sex chromosomes. The mitochondrial genome is a circular DNA molecule distinct from nuclear DNA. Although the mitochondrial DNA is very small compared to nuclear chromosomes, it codes for 13 proteins involved in mitochondrial energy production and specific tRNAs.
Foreign genetic material (most commonly DNA) can also be artificially introduced into the cell by a process called transfection. This can be transient, if the DNA is not inserted into the cell's genome, or stable, if it is. Certain viruses also insert their genetic material into the genome.
Organelles
Organelles are parts of the cell that are adapted and/or specialized for carrying out one or more vital functions, analogous to the organs of the human body (such as the heart, lung, and kidney, with each organ performing a different function). Both eukaryotic and prokaryotic cells have organelles, but prokaryotic organelles are generally simpler and are not membrane-bound.
There are several types of organelles in a cell. Some (such as the nucleus and Golgi apparatus) are typically solitary, while others (such as mitochondria, chloroplasts, peroxisomes and lysosomes) can be numerous (hundreds to thousands). The cytosol is the gelatinous fluid that fills the cell and surrounds the organelles.
Eukaryotic

- Cell nucleus: A cell's information center, the cell nucleus is the most conspicuous organelle found in a eukaryotic cell. It houses the cell's chromosomes, and is the place where almost all DNA replication and RNA synthesis (transcription) occur. The nucleus is spherical and separated from the cytoplasm by a double membrane called the nuclear envelope, space between these two membrane is called perinuclear space. The nuclear envelope isolates and protects a cell's DNA from various molecules that could accidentally damage its structure or interfere with its processing. During processing, DNA is transcribed, or copied into a special RNA, called messenger RNA (mRNA). This mRNA is then transported out of the nucleus, where it is translated into a specific protein molecule. The nucleolus is a specialized region within the nucleus where ribosome subunits are assembled. In prokaryotes, DNA processing takes place in the cytoplasm.
- Mitochondria and chloroplasts: generate energy for the cell. Mitochondria are self-replicating double membrane-bound organelles that occur in various numbers, shapes, and sizes in the cytoplasm of all eukaryotic cells. Respiration occurs in the cell mitochondria, which generate the cell's energy by oxidative phosphorylation, using oxygen to release energy stored in cellular nutrients (typically pertaining to glucose) to generate ATP (aerobic respiration). Mitochondria multiply by binary fission, like prokaryotes. Chloroplasts can only be found in plants and algae, and they capture the sun's energy to make carbohydrates through photosynthesis.

- Endoplasmic reticulum: The endoplasmic reticulum (ER) is a transport network for molecules targeted for certain modifications and specific destinations, as compared to molecules that float freely in the cytoplasm. The ER has two forms: the rough ER, which has ribosomes on its surface that secrete proteins into the ER, and the smooth ER, which lacks ribosomes. The smooth ER plays a role in calcium sequestration and release and also helps in synthesis of lipid.
- Golgi apparatus: The primary function of the Golgi apparatus is to process and package the macromolecules such as proteins and lipids that are synthesized by the cell.
- Lysosomes and peroxisomes: Lysosomes contain digestive enzymes (acid hydrolases). They digest excess or worn-out organelles, food particles, and engulfed viruses or bacteria. Peroxisomes have enzymes that rid the cell of toxic peroxides, Lysosomes are optimally active in an acidic environment. The cell could not house these destructive enzymes if they were not contained in a membrane-bound system.
- Centrosome: the cytoskeleton organizer: The centrosome produces the microtubules of a cell—a key component of the cytoskeleton. It directs the transport through the ER and the Golgi apparatus. Centrosomes are composed of two centrioles which lie perpendicular to each other in which each has an organization like a cartwheel, which separate during cell division and help in the formation of the mitotic spindle. A single centrosome is present in the animal cells. They are also found in some fungi and algae cells.
- Vacuoles: Vacuoles sequester waste products and in plant cells store water. They are often described as liquid filled spaces and are surrounded by a membrane. Some cells, most notably Amoeba, have contractile vacuoles, which can pump water out of the cell if there is too much water. The vacuoles of plant cells and fungal cells are usually larger than those of animal cells. Vacuoles of plant cells are surrounded by a membrane which transports ions against concentration gradients.
Eukaryotic and prokaryotic
- Ribosomes: The ribosome is a large complex of RNA and protein molecules. They each consist of two subunits, and act as an assembly line where RNA from the nucleus is used to synthesise proteins from amino acids. Ribosomes can be found either floating freely or bound to a membrane (the rough endoplasmatic reticulum in eukaryotes, or the cell membrane in prokaryotes).
- Plastids: Plastid are membrane-bound organelle generally found in plant cells and euglenoids and contain specific pigments, thus affecting the colour of the plant and organism. And these pigments also helps in food storage and tapping of light energy. There are three types of plastids based upon the specific pigments. Chloroplasts contain chlorophyll and some carotenoid pigments which helps in the tapping of light energy during photosynthesis. Chromoplasts contain fat-soluble carotenoid pigments like orange carotene and yellow xanthophylls which helps in synthesis and storage. Leucoplasts are non-pigmented plastids and helps in storage of nutrients.
Structures outside the cell membrane
Many cells also have structures which exist wholly or partially outside the cell membrane. These structures are notable because they are not protected from the external environment by the cell membrane. In order to assemble these structures, their components must be carried across the cell membrane by export processes.
Cell wall
Many types of prokaryotic and eukaryotic cells have a cell wall. The cell wall acts to protect the cell mechanically and chemically from its environment, and is an additional layer of protection to the cell membrane. Different types of cell have cell walls made up of different materials; plant cell walls are primarily made up of cellulose, fungi cell walls are made up of chitin and bacteria cell walls are made up of peptidoglycan.
Prokaryotic
Capsule
A gelatinous capsule is present in some bacteria outside the cell membrane and cell wall. The capsule may be polysaccharide as in pneumococci, meningococci or polypeptide as Bacillus anthracis or hyaluronic acid as in streptococci. Capsules are not marked by normal staining protocols and can be detected by India ink or methyl blue, which allows for higher contrast between the cells for observation.
Flagella
Flagella are organelles for cellular mobility. The bacterial flagellum stretches from cytoplasm through the cell membrane(s) and extrudes through the cell wall. They are long and thick thread-like appendages, protein in nature. A different type of flagellum is found in archaea and a different type is found in eukaryotes.
Fimbriae
A fimbria (plural fimbriae also known as a pilus, plural pili) is a short, thin, hair-like filament found on the surface of bacteria. Fimbriae are formed of a protein called pilin (antigenic) and are responsible for the attachment of bacteria to specific receptors on human cells (cell adhesion). There are special types of pili involved in bacterial conjugation.
Cellular processes

Replication
Cell division involves a single cell (called a mother cell) dividing into two daughter cells. This leads to growth in multicellular organisms (the growth of tissue) and to procreation (vegetative reproduction) in unicellular organisms. Prokaryotic cells divide by binary fission, while eukaryotic cells usually undergo a process of nuclear division, called mitosis, followed by division of the cell, called cytokinesis. A diploid cell may also undergo meiosis to produce haploid cells, usually four. Haploid cells serve as gametes in multicellular organisms, fusing to form new diploid cells.
DNA replication, or the process of duplicating a cell's genome, always happens when a cell divides through mitosis or binary fission. This occurs during the S phase of the cell cycle.
In meiosis, the DNA is replicated only once, while the cell divides twice. DNA replication only occurs before meiosis I. DNA replication does not occur when the cells divide the second time, in meiosis II. Replication, like all cellular activities, requires specialized proteins for carrying out the job.
DNA repair
Cells of all organisms contain enzyme systems that scan their DNA for damage and carry out repair processes when it is detected. Diverse repair processes have evolved in organisms ranging from bacteria to humans. The widespread prevalence of these repair processes indicates the importance of maintaining cellular DNA in an undamaged state in order to avoid cell death or errors of replication due to damage that could lead to mutation. E. coli bacteria are a well-studied example of a cellular organism with diverse well-defined DNA repair processes. These include: nucleotide excision repair, DNA mismatch repair, non-homologous end joining of double-strand breaks, recombinational repair and light-dependent repair (photoreactivation).
Growth and metabolism
Between successive cell divisions, cells grow through the functioning of cellular metabolism. Cell metabolism is the process by which individual cells process nutrient molecules. Metabolism has two distinct divisions: catabolism, in which the cell breaks down complex molecules to produce energy and reducing power, and anabolism, in which the cell uses energy and reducing power to construct complex molecules and perform other biological functions.
Complex sugars can be broken down into simpler sugar molecules called monosaccharides such as glucose. Once inside the cell, glucose is broken down to make adenosine triphosphate (ATP), a molecule that possesses readily available energy, through two different pathways. In plant cells, chloroplasts create sugars by photosynthesis, using the energy of light to join molecules of water and carbon dioxide.
Protein synthesis
Cells are capable of synthesizing new proteins, which are essential for the modulation and maintenance of cellular activities. This process involves the formation of new protein molecules from amino acid building blocks based on information encoded in DNA/RNA. Protein synthesis generally consists of two major steps: transcription and translation.
Transcription is the process where genetic information in DNA is used to produce a complementary RNA strand. This RNA strand is then processed to give messenger RNA (mRNA), which is free to migrate through the cell. mRNA molecules bind to protein-RNA complexes called ribosomes located in the cytosol, where they are translated into polypeptide sequences. The ribosome mediates the formation of a polypeptide sequence based on the mRNA sequence. The mRNA sequence directly relates to the polypeptide sequence by binding to transfer RNA (tRNA) adapter molecules in binding pockets within the ribosome. The new polypeptide then folds into a functional three-dimensional protein molecule.
Motility
Unicellular organisms can move in order to find food or escape predators. Common mechanisms of motion include flagella and cilia.
In multicellular organisms, cells can move during processes such as wound healing, the immune response and cancer metastasis. For example, in wound healing in animals, white blood cells move to the wound site to kill the microorganisms that cause infection. Cell motility involves many receptors, crosslinking, bundling, binding, adhesion, motor and other proteins. The process is divided into three steps: protrusion of the leading edge of the cell, adhesion of the leading edge and de-adhesion at the cell body and rear, and cytoskeletal contraction to pull the cell forward. Each step is driven by physical forces generated by unique segments of the cytoskeleton.
Navigation, control and communication
In August 2020, scientists described one way cells—in particular cells of a slime mold and mouse pancreatic cancer-derived cells—are able to navigate efficiently through a body and identify the best routes through complex mazes: generating gradients after breaking down diffused chemoattractants which enable them to sense upcoming maze junctions before reaching them, including around corners.
Multicellularity
Cell specialization/differentiation
Multicellular organisms are organisms that consist of more than one cell, in contrast to single-celled organisms.
In complex multicellular organisms, cells specialize into different cell types that are adapted to particular functions. In mammals, major cell types include skin cells, muscle cells, neurons, blood cells, fibroblasts, stem cells, and others. Cell types differ both in appearance and function, yet are genetically identical. Cells are able to be of the same genotype but of different cell type due to the differential expression of the genes they contain.
Most distinct cell types arise from a single totipotent cell, called a zygote, that differentiates into hundreds of different cell types during the course of development. Differentiation of cells is driven by different environmental cues (such as cell–cell interaction) and intrinsic differences (such as those caused by the uneven distribution of molecules during division).
Origin of multicellularity
Multicellularity has evolved independently at least 25 times, including in some prokaryotes, like cyanobacteria, myxobacteria, actinomycetes, or Methanosarcina. However, complex multicellular organisms evolved only in six eukaryotic groups: animals, fungi, brown algae, red algae, green algae, and plants. It evolved repeatedly for plants (Chloroplastida), once or twice for animals, once for brown algae, and perhaps several times for fungi, slime molds, and red algae. Multicellularity may have evolved from colonies of interdependent organisms, from cellularization, or from organisms in symbiotic relationships.
The first evidence of multicellularity is from cyanobacteria-like organisms that lived between 3 and 3.5 billion years ago. Other early fossils of multicellular organisms include the contested Grypania spiralis and the fossils of the black shales of the Palaeoproterozoic Francevillian Group Fossil B Formation in Gabon.
The evolution of multicellularity from unicellular ancestors has been replicated in the laboratory, in evolution experiments using predation as the selective pressure.
Origins
The origin of cells has to do with the origin of life, which began the history of life on Earth.
Origin of life

Small molecules needed for life may have been carried to Earth on meteorites, created at deep-sea vents, or synthesized by lightning in a reducing atmosphere. There is little experimental data defining what the first self-replicating forms were. RNA may have been the earliest self-replicating molecule, as it can both store genetic information and catalyze chemical reactions.
Cells emerged around 4 billion years ago. The first cells were most likely heterotrophs. The early cell membranes were probably simpler and more permeable than modern ones, with only a single fatty acid chain per lipid. Lipids spontaneously form bilayered vesicles in water, and could have preceded RNA.
First eukaryotic cells
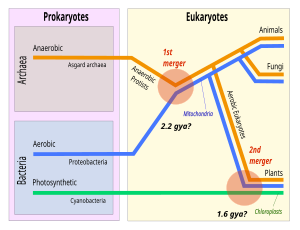
Eukaryotic cells were created some 2.2 billion years ago in a process called eukaryogenesis. This is widely agreed to have involved symbiogenesis, in which archaea and bacteria came together to create the first eukaryotic common ancestor. This cell had a new level of complexity and capability, with a nucleus and facultatively aerobic mitochondria. It evolved some 2 billion years ago into a population of single-celled organisms that included the last eukaryotic common ancestor, gaining capabilities along the way, though the sequence of the steps involved has been disputed, and may not have started with symbiogenesis. It featured at least one centriole and cilium, sex (meiosis and syngamy), peroxisomes, and a dormant cyst with a cell wall of chitin and/or cellulose. In turn, the last eukaryotic common ancestor gave rise to the eukaryotes' crown group, containing the ancestors of animals, fungi, plants, and a diverse range of single-celled organisms. The plants were created around 1.6 billion years ago with a second episode of symbiogenesis that added chloroplasts, derived from cyanobacteria.
History of research

In 1665, Robert Hooke examined a thin slice of cork under his microscope, and saw a structure of small enclosures. He wrote "I could exceeding plainly perceive it to be all perforated and porous, much like a Honey-comb, but that the pores of it were not regular". To further support his theory, Matthias Schleiden and Theodor Schwann both also studied cells of both animal and plants. What they discovered were significant differences between the two types of cells. This put forth the idea that cells were not only fundamental to plants, but animals as well.
- 1632–1723: Antonie van Leeuwenhoek taught himself to make lenses, constructed basic optical microscopes and drew protozoa, such as Vorticella from rain water, and bacteria from his own mouth.
- 1665: Robert Hooke discovered cells in cork, then in living plant tissue using an early compound microscope. He coined the term cell (from Latin cellula, meaning "small room") in his book Micrographia (1665).
- 1839: Theodor Schwann and Matthias Jakob Schleiden elucidated the principle that plants and animals are made of cells, concluding that cells are a common unit of structure and development, and thus founding the cell theory.
- 1855: Rudolf Virchow stated that new cells come from pre-existing cells by cell division (omnis cellula ex cellula).
- 1931: Ernst Ruska built the first transmission electron microscope (TEM) at the University of Berlin. By 1935, he had built an EM with twice the resolution of a light microscope, revealing previously unresolvable organelles.
- 1981: Lynn Margulis published Symbiosis in Cell Evolution detailing how eukaryotic cells were created by symbiogenesis.