Epigenetics of anxiety and stress–related disorders is the field studying the relationship between epigenetic modifications of genes and anxiety and stress-related disorders, including mental health disorders such as generalized anxiety disorder (GAD), post-traumatic stress disorder, obsessive-compulsive disorder (OCD), and more. These changes can lead to transgenerational stress inheritance.
Epigenetic modifications play a role in the development and heritability of these disorders and related symptoms. For example, regulation of the hypothalamus-pituitary-adrenal axis by glucocorticoids plays a major role in stress response and is known to be epigenetically regulated.
As of 2015 most work has been done in animal models in laboratories, and little work has been done in humans; the work is not yet applicable to clinical psychiatry. Stress-induced epigenetic changes, particularly to genes that effect the hypothalamic–pituitary–adrenal (HPA) axis, persist into future generations, negatively impacting the capacity of offspring to adapt to stress. Early life experiences, even when generations removed, can cause permanent epigenetic modifications of DNA resulting in changes in gene expression, endocrine function and metabolism. These heritable epigenetic modifications include DNA methylation of the promoter regions of genes that affect sensitivity to stress.
Mechanism
Epigenetic modification in response to stress results in molecular and genetic alterations that in turn results in mis-regulated or silenced genes. Heterochromatin is the protein that controls the silencing of these genes epigenetically. For example, epigenetic modifications to the gene BDNF (brain derived neurotrophic factor), as well as Drosophila ATF-2 (dATF-2), as a result of stress can be passed on to offspring. Chronic variable stress induces offspring hypothalamic gene expression modifications, including elevated methylation levels of the BDNF promoter in the hippocampus. This methylation will also occur in the heterochromatin, causing a disrupted heterochromatin to be passed on to the child. Maternal separation and postnatal maternal abuse also increases DNA methylation at regulatory regions of BDNF genes in the prefrontal cortex and hippocampus, leading to potential stress vulnerability in future generations.
Stress can also result in inheritable changes DNA methylation in the promoter regions of the estrogen receptor alpha (ERα), glucocorticoid receptor (GR), and mineralocorticoid receptor (MR). These changes lead to altered expression of these genes in offspring that in turn leads to decreased stress tolerance.
Stress and the HPA axis
Gene regulation as it relates to the HPA axis has been implicated in transgenerational stress effects. Environmental prenatal stress exposure, for example, alters glucocorticoid receptor gene expression, gene function, and future stress response in F1 and F2 generations. Maternal care likewise contributes to HPA-related epigenetic modifications. Epigenetic re-programming of gene expression alters stress response in offspring later in life when exposed to decreased maternal care. Inattentive mothering has led to increased levels of gene methyl marks, compared to attentive mothers. Female offspring with low licking-grooming mothers have decreased promoter methylation and increased histone acetylation, leading to increased glucocorticoid receptor expression. Epigenetic modifications as a result of absent maternal care lead to decreased estrogen receptor alpha expression, due to increased methylation at the gene's promoter.
Epigenetic writers, erasers, and readers
Epigenetic changes are performed by enzymes known as writers, which can add epigenetic modifications, erasers, which erase epigenetic modifications, and readers, which can recognize epigenetic modifications and cause a downstream effect. Stress-induced modifications of these writers, erasers, and readers result in important epigenetic modifications such as DNA methylation and acetylation.
DNA methylation

DNA methylation is a type of epigenetic modification in which methyl groups are added to cytosines of DNA. It is located on the fifth position of cytosine which has importance in the development of mammals. DNA methylation is an important regulator of gene expression and is usually associated with gene repression. DNA Methylation is a mechanism which can suppress gene expression. It can be inherited through cell divisions in development, and is involved with cell memory. Changes in methylation occur due to mutated or deregulated chromatin regulators. This process is also used in marking cancers for diagnosis.
MeCP2
Laboratory studies have found that early life stress in rodents can cause phosphorylation of methyl CpG binding protein 2 (MeCP2), a protein that preferentially binds CpGs and is most often associated with suppression of gene expression. Stress-dependent phosphorylation of MeCP2 causes MeCP2 to dissociate from the promoter region of a gene called arginine vasopressin (avp), causing avp to become demethylated and upregulated. This may be significant because arginine vasopressin is known to regulate mood and cognitive behavior. Additionally, arginine vasopressin upregulates corticotropin-releasing hormone (CRH), which is a hormone important for stress response. Thus, stress-induced upregulation of avp due to demethylation might alter mood, behavior, and stress responses. Demethylation of this locus can be explained by reduced binding of DNA methyl transferases (DNMT), an enzyme that adds methyl groups to DNA, to this locus.
MeCP2 is known to have interactions with several other enzymes that modify chromatin (for example, HDAC-containing complexes and co-repressors) and in turn regulate activity of genes that modulate stress response either by increasing or decreasing stress tolerance. For example, epigenetic upregulation of genes that increase stress response may cause decreased stress tolerance in an organism. These interactions are dependent on the phosphorylation status of MeCP2, which as previously mentioned, can be altered by stress.
DNMT1
DNA methyltransferase 1 (DNMT1) belongs to a family of proteins known as DNA methyltransferases, which are enzymes that add methyl groups to DNA. DNMT1 is specifically involved in maintaining DNA methylation; hence it is also known as the maintenance methylase DNMT1. DNMT1 aids in regulation of gene expression by methylating promoter regions of genes, causing transcriptional repression of these genes.
DNMT1 is transcriptionally repressed under stress-mimicking exposure both in vitro and in vivo using a mouse model. Accordingly, transcriptional repression of DNMT1 in response to long-term stress-mimicking exposure causes decreased DNA methylation, which is a marker of gene activation. In particular, there is decreased methylation of a gene called fkbp5, which plays a role in stress response as a glucocorticoid-responsive gene. Thus, chronic stress may cause demethylation and hyperactivation of a stress-related gene, causing increased stress response.
Additionally, DNMT1 gene locus has increased methylation in individuals who were exposed to trauma and developed post-traumatic stress disorder (PTSD). Increased methylation of DNMT1 did not occur in trauma-exposed individuals who did not develop PTSD. This may indicate an epigenetic phenotype that can differentiate PTSD-susceptible and PTSD-resilient individuals after exposure to trauma.
Transcription factors
Transcription factors are proteins that bind DNA and modulate the transcription of genes into RNA such as mRNA, tRNA, rRNA, and more; thus they are essential components of gene activation. Stress and trauma can affect expression of transcription factors, which in turn alter DNA methylation patterns.
For example, transcription factor nerve growth-induced protein A (NGFI-A, also called NAB1) is up-regulated in response to high maternal care in rodents, and down-regulated in response to low maternal care (a form of early life stress). Decreased NGFI-A due to low maternal care increases methylation of a glucocorticoid receptor promoter in rats. Glucocorticoid is known to play a role in downregulating stress response; therefore, downregulation of glucocorticoid receptor by methylation causes increased sensitivity to stress.
Histone acetylation

Histone acetylation and deacetylation is a type of epigenetic modification in which acetyl groups are added to lysine on histone tails. Histone acetylation, performed by enzymes known as histone acetyltransferases (HATs), removes the positive charge from lysine and results in gene activation by weakening the histone's interaction with negatively-charged DNA. In contrast, histone deacetylation performed by histone deacetylases (HDACs) results in gene deactivation.
HDAC
Transcriptional activity and expression of HDACs is altered in response to early life stress. For animals exposed to early life stress, HDAC expression tends to be lower when they are young and higher when they are older. This suggests an age-dependent effect of early life stress on HDAC expression. These HDACs may result in deacetylation and thus activation of genes that upregulate stress response and decrease stress tolerance.
Transgenerational epigenetic influences
Genome-wide association studies have shown that psychiatric disorders are partly heritable; however, heritability cannot be fully explained by classical Mendelian genetics, but rather epigenetics. There are many components in understanding the heritability of psychiatric disorders. Understanding epigenetic modifications and its ability to impact epigenomes over generations is vital in analyzing potential behavioral disorders. But we must acknowledge the concept of transgenerational epigenetics (epigenetic inheritance) which is the occurrence in which parents are able to transfer traits not present in their DNA sequencing to their offspring; it is the passing of environmentally manipulated traits for two or more generations without direct DNA alteration. For example, one study found transmission of DNA methylation patterns from fathers to offspring during spermatogenesis. Similarly, several studies have shown that traits of psychiatric illnesses (such as traits of PTSD and other anxiety disorders) can be transmitted epigenetically Parental exposure to various stimuli, both positive and negative, can cause transgenerational epigenetic and behavioral effects.
Parental exposure to trauma and stress
Trauma and stress experienced by a parent can cause epigenetic changes to its offspring. This has been observed both in population and experimental studies.
Biological vulnerability and HPA axis alterations may be observed after maternal epigenetic programing during pregnancy, leading to similar modifications in future generations. Child abuse exposure, for example, is associated with lower baseline infant cortisol levels as well as modified HPA axis function. Human studies investigating posttraumatic stress disorder (PTSD) and its effects on offspring have illustrated similar molecular and HPA axis modification and function. PTSD patients who experienced trauma from genocides or terrorist attacks frequently exhibited aggressive or neglectful behavior toward offspring during critical developmental periods, possibly contributing to permanent glucocorticoid deregulation in offspring. PTSD mothers and children illustrate lower basal cortisol levels and glucocorticoid receptors and increased mineralocorticoid receptors when exposed to stress. Therefore, developmental experiences, such as stress exposure, may have critical effects on neuromodulatory mechanisms transgenerationally.
Strong relationships between maternal care and subsequent epigenetic modification in offspring, similar to that found in animal models, has been observed in humans. Severe emotional trauma in the mother, for example, often leads to modified methylation patterns of DNA in subsequent offspring generations. PTSD exposed offspring illustrate epigenetic modifications similar to that seen in PTSD mothers, with an increased NR3C2 methylation in exon 1 and increased CpG methylation in the NR3C2 coding sequence, leading to alterations in mineralocorticoid receptor gene expression. Additionally, investigation of post mortem hippocampal tissue indicates decreased levels of neuron-specific glucocorticoid receptor mRNA and decreased DNA methylation in promoter regions among suicidal individuals with lifelong stress or abuse exposure.
Epigenetic mechanisms as a result of early life stress may be responsible for neuronal and synaptic alterations in the brain. Developmental stress exposure has been shown to alter brain structure and behavioral functions in adulthood. Evidence of decreased complexity in the CA1 and CA3 region of the hippocampus in terms of dendritic length and spine density after early-life stress exposure indicates transgenerational stress inheritance. Therefore, environmental and experience-dependent synaptic reorganization and structure modifications may lead to increased stress vulnerability and brain dysfunction in future generations.
Transgenerational Stress Effects
Human models illustrating transgenerational stress effects are limited due to relatively novel exploration of the topic of epigenetics as well as lengthy follow-up intervals required for multi-generational studies. Several models, however, have investigated the role of epigenetic inheritance and transgenerational stress effects. Transgenerational stress in humans, as in animal models, induces effects influencing social behavior, reproductive success, cognitive ability, and stress response. Similar to animal models, human studies have investigated the role of epigenetics and transgenerational inheritance molecularly as it relates to the HPA system. Prenatal influences, such as emotional stress, nutrition deprivation, toxin exposure, hypoxia, increased maternal HPA activity, and cortisol levels may activate or affect HPA axis activity of offspring, despite placental barrier.
Paternal stress inheritance
Paternal stress is an important factor in the determination of inheritance of genes as well as maternal stress inheritance. Factors such as environment and experiences can alter the epigenetic of paternal genes as well as in sperm. Epigenetic changes to the DNA in sperm ("epigenetic tags") prior to conception can be passed to offspring. The paternal phenotype will be inherited into the offspring due to genetic information being stored in the sperm. In studies, it is shown as rodent offspring are fostered mono-parentally and have no direct exposure with their fathers, offspring born of stressed male rodents provide a good model for transgenerational stress inheritance. Direct injection of sperm RNAs to wild type oocytes results in reproducible stress-related modifications. Small non-coding RNAs may serve as a potential mechanism for stress-related genetic changes in offspring. Mouse models of traumatic early life stress exposure result in microRNA modifications and subsequent differences in gene expression and metabolic function. This effect was reproducible by sperm RNA injection, leading to similar gene modifications in future generations. The novelty of this research suggests direct mechanisms capable of altering epigenetics by stress-related factors.
Phenotypic effects
Early life experiences and environmental factors may lead to epigenetic modification at specific gene loci, leading to altered neuronal plasticity, function, and subsequent behavior. As mentioned, there are genetic markers in all living organisms. There can also be the presence of epigenetically marks and this are basically areas of modification on DNA that in with gene expression. Certain exposures within the environment can lead to the expression of genes in various ways which can contribute to behavioral plasticity patterns that can potentially also change the ways in which organism functions when under normal conditions. Chromatin remodeling in rodent offspring and altered gene expression within the limbic brain regions that may contribute to depression, stress, and anxiety-related disorders in future generations. Variations in maternal care, such as maternal licking and grooming, indicates reduced HPA axis reactivity in subsequent generations. Such HPA axis modifications lead to decreased anxiety-like behavior in adulthood and increased glucocorticoid receptor levels leading to negative feedback on HPA reactivity and further behavioral modifications. Rodent models of maternal separation also reveal increased depressive-like behavior in offspring, decreased stress coping abilities, and changes in DNA methylation.

Holocaust
An epidemiological study investigating behavioral, physiological, and molecular changes in the children of Holocaust survivors found epigenetic modifications of a glucocorticoid receptor gene, Nr3c1. This is significant because glucocorticoid is a regulator of the hypothalamus-pituitary-adrenal axis (HPA) and is known to affect stress response. These stress-related epigenetic changes were accompanied by other characteristics that indicated higher stress and anxiety in these offspring, including increased symptoms of PTSD, greater risk of anxiety, and higher levels of the stress hormone cortisol. The offspring demonstrate greater risk of developing PTSD in response to their own trauma or traumas. Offspring with maternal exposure to the Holocaust during the mother's childhood has demonstrated significantly lower site 6 methylation. The site 6 methylation impacts the stress response. In addition to PTSD risks in response to individual trauma in offspring, there has also been an increase in nightmares of offspring related to persecution and torment.
Experimental evidence
The effect of parental exposure to stress has been tested experimentally as well. For example, male mice who were put under early life stress through poor maternal care—a scenario analogous to human childhood trauma—passed on epigenetic changes that resulted in behavioral changes in offspring. Offspring experienced altered DNA methylation of stress-response genes such as CB1 and CRF2 in the cortex, as well as epigenetic alterations in transcriptional regulation gene MeCP2. Offspring were also more sensitive to stress, which is in accordance with the altered epigenetic profile. These changes persisted for up to three generations.
In another example, male mice were socially isolated as a form of stress. Offspring of these mice had increased anxiety in response to stressful conditions, increased stress hormone levels, dysregulation of the HPA axis which plays a key role in stress response, and several other characteristics that indicated increased sensitivity to stress.
Inheritance of small-noncoding RNA
Studies have found that early life stress induced through poor maternal care alters sperm epigenome in male mice. In particular, expression patterns of small-noncoding RNAs (sncRNAs) are altered in the sperm, as well as in stress-related regions of the brain. Offspring of these mice exhibited the same sncRNA expression changes in the brain, but not in the sperm. These changes were coupled with behavioral changes in the offspring that were comparable to behavior of the stressed fathers, especially in terms of stress response. Additionally, when the sncRNAs in the fathers' sperm were isolated and injected into fertilized eggs, the resulting offspring inherited the stress behavior of the father. This suggests that stress-induced modifications of sncRNAs in sperm can cause inheritance of stress phenotype independent of the father's DNA.
Parental exposure to positive stimulation
Exercise
Just as parental stress can alter epigenetics of offspring, parental exposure to positive environmental factors cause epigenetic modifications as well. For example, male mice that participated in voluntary physical exercise resulted in offspring that had reduced fear memory and anxiety-like behavior in response to stress. This behavioral change likely occurred due to expressions of small non-coding RNAs that were altered in sperm cells of the fathers. Participation in aerobic exercise led to decreased cortisol levels in males.
Stress effect reversal
Additionally, exposing fathers to enriching environments can reverse the effect of early life stress on their offspring. When early life stress is followed by environmental enrichment, anxiety-like behavior in offspring is prevented. Similar studies have been conducted in humans and suggest that DNA methylation plays a role. Other studies have been conducted to find drugs such as T2D and PPArG can be used as an epigenetic regulation for tissues associated with diabetes. These drugs used show evidence for the therapies that can be associated with the stress effect reversal.
Childhood exposure to trauma
Early life development and childhood trauma

Healthy development early in life is critical. Early life is characterized by rapid development and increased susceptibility to modifications. Childhood trauma can severely affect the development of the brain, resulting in the alteration of neural circuits which are involved in emotional regulation and threat detection. Childhood trauma has been associated with a wide array of mental health disorders such as bipolar disorder, anxiety, post traumatic stress disorder (PTSD), and depression.
PTSD Inheritability
Research involving PTSD in those who experienced childhood trauma had a 25% to 60% inheritability rate, which is a relatively low to moderate rate. This study suggests that other factors play a role in the contribution to this disorder such as gene interactions involving epigenetic modifications. These epigenetic modifications, specifically DNA methylation can lead to the phenotypic expression of mental disorders.
Changes to the HPA Axis Due to Childhood Trauma
Hypothalamic-pituatary-adrenal (HPA) axis is essential component of the neuroendocrine system that regulates stress response. Persistent dysregulation of the stress response pathway resulting from childhood trauma causes alterations in the (HPA). These alterations lead to prolonged harmful physiological and physical changes.
Postmortem Brain Tissue DNA Methylation
A DNA methylation study was done by Labonte on postmortem human brain tissue comparing humans with or without a history of childhood abuse who died by suicide. Childhood abuse and trauma was associated with increased cytosine methylation of the NR3C1 promoter resulting in the decrease of GR expression. The NR3C1 gene encodes glucocorticoid receptor (GR) which is essential for glucose regulation and managing stress response through both genetic and epigenetic pathways.
Post-traumatic stress disorder (PTSD)
Post-traumatic stress disorder (PTSD) is an stress-related mental health disorder that emerges in response to traumatic or highly stressful experiences. It is believed that PTSD develops as a result of an interaction between these traumatic experiences and genetic factors. The signs and symptoms of PTSD can include avoidance behaviors, invasive thoughts, and significant alterations in normal behavior and thinking. There is evidence suggesting PTSD formation is associated with epigenetic changes such as DNA methylation and acetylation of histone proteins. Increased DNA methylation has been found to regulate the induction of fear conditioning behaviors associated with PTSD triggers. Histone modifications, like acetylation and deacetylation, play an important role in the development of PTSD, which is related to fear memory from traumatic events.
The DSM-5 asserts that PTSD manifests differently in children over six years old than in adults. Specifically that their flashbacks or intrusive memories may be explained by recreating their traumatic event(s) through their play. They may also experience reoccurring nightmares that are indirectly related to the event. Additionally, there is a separate criteria altogether for PTSD in children under six years old.
Epigenetic modifications
DNA methylation

Through a number of human studies, PTSD is known to affect DNA methylation of CpG islands in several genes involved in numerous activities, including stress responses and neurotransmitter activity. CpGs are used to describe cytosine-guanine adjacent nucleotides within the same strand of DNA. CpG islands are defined by computer algorithms as being made up of at least 60% CpGs and being anywhere between 200 and 3000 base pairs in size. The methylation of these CpG islands can cause histone modifications which can lead to the condensation of chromatin which can ultimately alter gene expression.
DNMT enzyme
DNA methyltransferase, DNMT, is an enzyme responsible for increased methylation of DNA. It has been found that DNMT and its associated increased methylation can regulate risk for memory consolidation and fear conditioning.
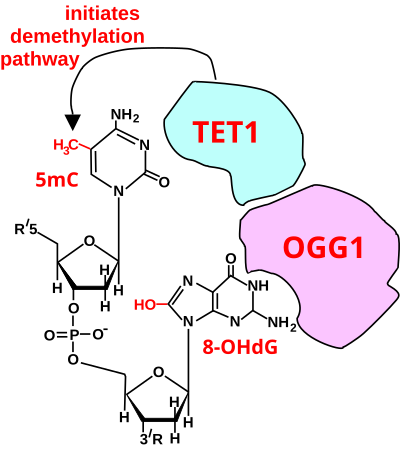
TET enzyme
The removal of methyl groups from cytosine is initiated by a TET enzyme. TET is an enzyme known to oxidize 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) within the genome. This reaction initiates active DNA demethylation to ultimately alter gene expression. It has been found that the TET enzyme exists as two isoforms which are differentially regulated and expressed across brain regions. The regulation of these isoforms can affect synaptic connections and ultimately memory formation. The manipulation of the TET enzymes' expression levels has become a potential source of interest for PTSD medication.
The table below identifies differentially methylated regions (DMRs) across the genome which undergo PTSD-induced epigenetic changes which alter gene expression.
| Genetic Loci | Finding(s) |
|---|---|
| SLC6A4 | Following trauma exposure, low methylation levels of SLC6A4 increases risk of PTSD; high methylation levels decreases risk of PTSD |
| MAN2C1 | Higher MAN2C1 methylation is correlated to greater risk of PTSD in individuals exposed to traumatic events |
| TPR, CLEC9A, APC5, ANXA2, TLR8 | PTSD is associated with increased global methylation of these genes |
| ADCYAP1R1 | Higher methylation is associated with PTSD symptoms in individuals exposed to trauma |
| LINE-1, Alu | Higher methylation of these loci is observed in postdeployed veterans who developed PTSD compared to those who do not develop PTSD |
| SLC6A3 | High SLC6A3 promoter methylation, combined with a nine-repeat allele of SLC6A3, is correlated to higher PTSD risk |
| IGF2, H19, IL8, IL16, IL18 | Higher methylation of IL18 but lower methylation of H19 and IL18 is associated with deployed veterans who developed PTSD |
| NR3C1 | Lower methylation levels of NR3C1 1B and 1C promoters is associated with PTSD;
Fathers with PTSD have offspring with higher NR3C1 1F promoter methylation; Lower levels of NR3C1 1F promoter methylation is associated with PTSD in combat veterans; Higher levels of NR3C1 methylation in male (but not female) Rwandan genocide survivors is associated with decreased PTSD risk |
| SPATC1L | Higher methylation is associated with PTSD symptoms. |
| HLA-DPB1 | Higher methylation is associated with PTSD symptoms. |
Histone modifications
Histone acetylation is performed by histone acetyl transferases (HATs) and histone deacetylation is carried out by histone deacetylases (HDACs). In rodent PTSD models, it has been found that an increase in histone acetylation is associated with fear conditioning. Histone acetylation can be involved in all parts of fear memory, including the development to memory extinction. It can also play a role in long-term potentiation (LTP). It was also observed that HDACs increase memory formation in fear extinction and HDAC inhibitors (HDACi) have shown evidence for modifying memory extinction, a possible treatment for PTSD.
Nervous system structures affected by PTSD
Hypothalamus-pituitary-adrenal axis
The hypothalamus-pituitary-adrenal (HPA) axis is a neuroendocrine system largely involved in ascertaining the levels of cortisol circulating the body at any given point in time. As cortisol plays a key role in the stress response, so does the HPA axis. The dysregulation of the HPA axis has been found to be characteristic of several stress disorders, including PTSD. This system works under a negative feedback loop structure. Hence, this HPA axis dysregulation may take the form of amplified negative inhibition and result in down-regulated cortisol levels. Epigenetic modifications play a role in this dysregulation, and these modifications are likely caused by the traumatic/stressful experience that triggered PTSD.
Immune dysregulation by HPA axis modifications
PTSD is often linked with immune dysregulation. Traumatic experiences can induce epigenetic changes at the gene loci that are immune-related which can lead to immune dysregulation and an increased risk of PTSD. Trauma exposure can also disrupt the HPA axis, thus altering peripheral immune function. The effect of PTSD on immune function arises in at least two ways: 1) Continuous disturbances on the HPA axis can dysregulate peripheral immune function, and 2) the effects of immune dysregulation in the periphery can lead to increased development of PTSD because of alterations in brain function.
PTSD-associated changes in immune cells found in blood or saliva can serve as biomarkers that trigger epigenetic changes which are involved in the pathogenesis of PTSD. These unique biomarkers serve as means of identifying PTSD subtypes. Beyond identifying subtypes, these distinct biomarkers can potentially be used to develop PTSD treatments.
Epigenetic modifications have been observed in immune-related genes of individuals with PTSD. For example, deployed military members who developed PTSD have higher methylation in the immune-related gene interleukin-18 (IL-18). This has interested scientists because high levels of IL-18 increase cardiovascular disease risk, and individuals with PTSD have elevated cardiovascular disease risk. Thus, stress-induced immune dysregulation via methylation of IL-18 may play a role in cardiovascular disease in individuals with PTSD.
Additionally, an epigenome-wide study found that individuals with PTSD have altered levels of methylation in the following immune-related genes: TPR, CLEC9A, APC5, ANXA2, TLR8, IL-4, and IL-2. This again shows that immune function in PTSD is disrupted, especially by epigenetic changes that are likely stress-induced.
Genes affected by PTSD
NR3C1
Nr3c1 is a transcription factor that encodes a glucocorticoid receptor (GR) and contains many GR response elements. Npas4 is another regulatory transcription factor also responsible for the regulation of GRs. Stress-induced changes in Nr3c1 and Npas4 methylation have been shown to alter stress sensitivity. This response differs between short-lived stress exposure and chronic stress exposure. In response to short lived stress, the NR3C1 promoter is more hydroxymethylated which is a modification associated with increased transcription of GR-associated genes. Thus, short lived stress exposure increases stress sensitivity. Conversely, in response to chronic stress, the Npas4 promoter has been presumed to be increased in methylation, a modification which is associated with inhibitory regulation of GRs. Thus, chronic stress exposure decreases stress sensitivity. These distinctions are important in understanding the epigenetic patterns of stress and genetic interactions with PTSD triggers. Overall, in the hippocampus of chronically stressed animals, the 3′-UTR (untranslated region of DNA) of the glucocorticoid receptor Nr3c1 showed increased hydroxymethylation, which led to increased transcription and thus, the disruption of stress tolerance and increased risk of disorders such as PTSD. However, early life stress increases methylation of the 1F promoter in this gene (or the 17 promoter analog in rodents). Because of its role in stress response and its link to early life stress, this gene has been of particular interest in the context of PTSD and has been studied in PTSD of both combat veterans and civilians.
In studies involving combat veterans, those who developed PTSD had lowered methylation of the Nr3c1 1F promoter compared to those who did not develop PTSD. Additionally, veterans who developed PTSD and had higher Nr3c1 promoter methylation responded better to long-term psychotherapy compared to veterans with PTSD who had lower methylation. These findings were recapitulated in studies involving civilians with PTSD. In civilians, PTSD is linked to lower methylation levels in the T-cells of exons 1B and 1C of Nr3c1, as well as higher GR expression. Thus, it seems that PTSD causes lowered methylation levels of GR loci and increased GR expression. The methylation of GR in T-cells are investigated because of its role in regulating cell immunity which, as such, stores cellular memory with environmental factors. T-cell fragments from individual cell populations are preferred over homogenized tissue because of the drastic variation in DNA methylation patterns between different cell fragments.
Although these results of decreased methylation and hyperactivation of GR conflict with the effect of early life stress at the same loci, these results match previous findings that distinguish HPA activity in early life stress versus PTSD. For example, cortisol levels of HPA in response to early life stress is hyperactive, whereas it is hypoactive in PTSD. Thus, the timing of trauma and stress—whether early or later in life—can cause differing effects on HPA and GR.
FKBP5
Fkbp5 encodes a GR-responsive protein known as Fk506 binding protein 51 (FKBP5). FKBP5 is induced by GR activation and functions in negative feedback by binding GR and reducing GR signaling. There is particular interest in this gene because some FKBP5 alleles have been correlated with increased risk of PTSD and development of PTSD symptoms—especially in PTSD caused by early life adversity. Therefore, FKBP5 likely plays an important role in PTSD.
As mentioned previously, certain FKBP5 alleles are correlated to increase PTSD risk, especially due to early life trauma. It is now known that epigenetic regulation of these alleles is also an important factor. For example, CpG sites in intron 7 of FKBP5 are demethylated after exposure to childhood trauma, but not adult trauma. Additionally, methylation of FKBP5 is alters in response to PTSD treatment; thus methylation levels of FKBP5 might correspond to PTSD disease progression and recovery.
ADCYAP1 and ADCYAP1R1
Pituitary adenylate cyclase-activating polypeptide (ADCYAP1) and its receptor (ADCYAP1R1) are stress responsive genes that play a role in modulating stress, among many other functions. Additionally, high levels of ADCYAP1 in peripheral blood is correlated to PTSD diagnosis in females who have experienced trauma, thus making ADCYAP1 a gene of interest in the context of PTSD.
Epigenetic regulation of these loci in relation to PTSD still require further investigation, but one study has found that high methylation levels of CpG islands in ADCYAP1R1 can predict PTSD symptoms in both males and females.
Alcohol use disorder
Alcohol use disorder is a type of brain disorder that requires one to have dependency on alcohol. Alcohol use disorder can vary in severity. Alcohol dependence can impact stress and other disorders in many ways. For example, stress-related disorders such as anxiety and PTSD are known to increase risk of alcohol use disorder (AUD), and they are often co-morbid. Mental disorders that pair with AUD can impacts the brain in many ways. For example, AUD can have the ability to aid those diagnosed with depression by alleviating depression symptoms such as insomnia, restlessness, and/ or the ability to re-engage in normal activities, and re-engage in hobbies. Bipolar disorder can cause manic episodes ranging fro different sudden mood changes. AUD can also be used to stall the same symptoms expressed in depression as well as with bipolar disorder. In some cases AUD can cause other brain disorders to worsen itself, or the symptoms of the disorder. An example of this can be seen in some with Obessive-Compulsive Disorder (can typically include anxiety “triggers” that often cause an individual to have very specific compulsions or obsessions). With this type of disorder, although it can help in ways by relieving symptomatic stress, it can also aid in promoting addiction to alcohol which can be a negative impact if uncontrolled. This may in part be due to the fact that alcohol can alleviate some symptoms of these disorders, thus promoting dependence on alcohol. Conversely, early exposure to alcohol can increase vulnerability to stress and stress-related disorders. AUD is a type of epigenetic influencing disorder, it is able to be passed down generation to generation epigenetically following a process mentioned before as transgenerational epigenetics. Moreover, alcohol dependence and stress are known to follow similar neuronal pathways, and these pathways are often unable to be regulated by similar epigenetic modifications.
Histone acetylation
HDAC
Histone acetylation is dysregulated by alcohol exposure and dependence, often through dysregulated expression and activity of HDACs, which modulate histone acetylation by removing acetyl groups from lysines of histone tails. For example, HDAC expression is upregulated in chronic alcohol use models. Monocyte-derived dendritic cells of alcohol users have increased HDAC gene expression compared to non-users. These results are also supported by in vivo rat studies, which show that HDAC expression is higher in alcohol-dependent mice that in non-dependent mice. Furthermore, knockout of HDAC2 in mice helps lower alcohol dependence behaviors. The same pattern of HDAC expression is seen in alcohol withdrawal, but acute alcohol exposure has the opposite effect; in vivo, HDAC expression and histone acetylation markers are decreased in the amygdala.
Dysregulation of HDACs is significant because it can cause upregulation or downregulation of genes that have important downstream effects both in alcohol dependence and anxiety-like behaviors, and the interaction between the two. A key example is BDNF (see "BDNF" below).
BDNF
Brain-derived neurotrophic factor (BDNF) is a key protein that is dysregulated by HDAC dysregulation. BDNF is a protein that regulates the structure and function of neuronal synapses. It plays an important role in neuronal activation, synaptic plasticity, and dendritic morphology—all of which are factors that may affect cognitive function. Dysregulation of BDNF is seen both in stress-related disorders and alcoholism; thus BDNF is likely an important molecule in the interaction between stress and alcoholism.
For example, BDNF is dysregulated by acute ethanol exposure. Acute ethanol exposure causes phosphorylation of CREB, which can cause increased histone acetylation at BDNF loci. Histone acetylation upregulates BDNF, in turn upregulating a downstream BDNF target called activity-regulated cytoskeleton associated protein (Arc), which is a protein responsible for dendritic spine structure and formation. This is significant because activation of Arc can be associated with anxiolytic (anxiety-reducing) effects. Therefore, ethanol consumption can cause epigenetic changes that alleviate stress and anxiety, thereby creating a pattern of stress-induced alcohol dependence.
Alcohol dependence is exacerbated by ethanol withdrawal. This is because ethanol withdrawal has the opposite effect of ethanol exposure; it causes lowered CREB phosphorylation, lowered acetylation, downregulation of BDNF, and increase in anxiety. Consequently, ethanol withdrawal reinforces desire for anxiolytic effects of ethanol exposure. Moreover, it is proposed that chronic ethanol exposure results in upregulation of HDAC activity, causing anxiety-like effects that can no longer be alleviated by acute ethanol exposure.
Potential epigenetic drug treatments
The most common treatments for anxiety disorders at the moment are benzodiazepines, Buspirone, and antidepressants. However, around one/third of patients with anxiety disorders do not respond well to the current anxiolytics, and many others have treatment-resistant anxiety disorders. Recent research surrounding DNA methylation changes in genes in genes encoding proteins associated with the HPA axis, histone modifications, and sncRNAs point to epigenetic drugs potentially being effective treatment methods for anxiety disorders.
HDACi
Histone deacetylase inhibitors (HDACi) fall into five different classes, not to be confused with the four different classes of HDACs. The five classes of HDACi consist of (I) hydroxamic acids, (II) short-chain fatty acids, (III) benzamides, (IV) cyclic tetrapeptides, and (V) sirtuin inhibitors. The three classes of HDACs are class I, consisting of HDAC1, HDAC2, HDAC3, HDAC8, class II, consisting of HDAC4, HDAC5, HDAC6, HDAC7, HDAC9, HDAC10, class III, consisting of NAD+-dependent HDACS, and class IV, consisting of HDAC11. While most HDACi inhibit only specific classes of HDACs, certain HDACi can act against all classes, making them pan-inhibitors.
HDACi are currently being researched as potential anxiolytics. At the moment, the mechanism of action of HDAC inhibitors in the treatment of anxiety disorders is not clear, as they affect several targets and have multiple pharmacological effects besides the inhibition of HDACs. However, they have been shown to cause DNA demethylation, possibly due to an increase in the levels of TET1, which is a demethylating enzyme. In the human peripheral cells of patients with anxiety disorders and in animal models of anxiety disorders, genes such as GAD1, NR3C1, BDNF, MAOA, HECA, and FKBP5 are shown to be hypermethylated. As such, the mechanism of action of HDACi in anxiety disorders could, in part, be potentially explained by the demethylation of those genes.
Valproate
Valproate is a drug that acts as an HDACi on class I and II HDACs. Six clinical trials surrounding its use as an anxiolytic have been performed so far. Five of the six trials were performed on patients with anxiety disorders, and one of the trials used healthy subjects with no anxiety disorders. Of the five trials performed on patients with anxiety disorders, three found that Valproate decreases panic disorder, one found that Valproate decreases social anxiety, and one found that Valproate reduces generalized anxiety. The trial performed on healthy subjects found that Valproate reduces anxiety and also acts as a nerve conduction inhibitor, which could be an explanation for some of its anxiety-reducing effects.
D-cycloserine, Trichostatin-a, Suberoylanilide hydroxamic acid, sodium butyrate, and valproic acid
Various preclinical drug trials using other HDAC inhibitors have also been performed, with most drugs targeting HDAC classes I and II and a select few targeting classes IV and III. The HDACi drug, d-cycloserine, was found to reduce fear in 129S1/SvImJ mice, which are mice that show poor extinction acquisition and recovery of fear-induced suppression of heart-rate variability, enlarged dendritic arbors in basolateral amygdala neurons, and functional abnormalities in cortico-amygdala circuitry that mediates fear extinction. Trichostatin-a was normalized BDNF and Arc expression in the central and the medial nucleus of the amygdala in rats experiencing alcohol withdrawal. Suberoylanilide hydroxamic acid significantly reversed anxiety-like behaviors and stress-induced gastrointestinal hypersensitivity and fecal pellet output. Anxiety-like and depression-like behaviors caused by immobilization stress or nicotine addiction were also reduced in mice treated with the HDACi sodium butyrate and valproic acid.
Lactate
Lactate, a metabolite that is naturally produced during exercise, was found to function as an HDAC II and III modulator in a pre-clinical trial. The trial was performed on C57BL/6 mice, which are mice that were exposed to chronic stress in the form of daily defeat by a CD-1 aggressive mouse. While control mice exhibited increased social avoidance, anxiety, and susceptibility to depression, mice that received lactate before each defeat demonstrated resilience to depression and stress and reduced social avoidance and anxiety. Lactate promoted this resilience by restoring regular hippocampal class I HDAC levels and activity.
sncRNA
Preliminary research has been done about therapy involving small non-coding RNAs, demonstrating that they can regulate epigenetic mechanisms of gene expression and could present as biomarkers for disease. One therapy option is for the sncRNAs in patients with anxiety disorders to be targeted for upregulation. Another option is to inhibit the miRNAs in order to reduce their effects, potentially using antisense oligonucleotides or antagomirs as inhibitors.
Hydrocortisone
The medication hydrocortisone is a synthetic form of cortisol, and is typically an anti inflammatory. In recent years, the administration of hydrocortisone has been tested as a possible preventative measure for the onset of PTSD symptoms. Ideally, it should be administered immediately after a traumatic event. The efficacy of hydrocortisone as a preventative intervention for PTSD has been confirmed by a meta-analysis of eight separate studies, and researchers believe the best results are obtained when hydrocortisone is administered within the first six hours of exposure to the traumatic event. At this time, however, no curative properties have been discovered. Hydrocortisone's potential operates on two bases: restoration of normal HPA axis functioning and interference with memory consolidation.
HPa axis homeostasis
Our standard understandings of PTSD may suggest elevated glucocorticoid levels during and directly following events of trauma. However, multiple studies have indicated that overall HPA axis activity and cortisol levels are depleted in the critical aftermath and extended period after trauma. Moreover, research has also indicated that an appropriate release of glucocorticoids following acute stress may restore homeostatic equilibrium of the HPA axis, thereby preventing gradual sensitization, which is responsible for persistent cortisol reduction and increased PTSD susceptibility. Thus, the appropriately-dosed administration of hydrocortisone promptly following the traumatic incident would normalize the HPA axis and potentially prevent PTSD onset.
Disruption of memory consolidation
In the absence of memory reactivation, hydrocortisone's effectiveness within a six-hour window supports the consolidation theory, which asserts that memory is labile even immediately after trauma. It is assumed that the medication is disrupting initial memory consolidation of the traumatic event. However, its exact mechanism within this context remains largely unknown.
Although trials have proven promising, there is much more research to be done. Further comprehensive studies are required amidst more diverse populations under different traumatic conditions in order to ascertain factors of optimal usage of the drug and clarify the PTSD subgroups hydrocortisone is beneficial to.
Rodent models used to study PTSD medication
Stress-enhanced fear learning (SEFL)
The observation of epigenetic modifications and their role in regulating fear learning is an active area of research. The use of stress-enhanced fear learning (SEFL) paradigms are important for forming preclinical models of PTSD because one is able to observe the epigenetic changes in rodents and PTSD associated changes in fear learning after stress exposure.
Single-prolonged stress (SPS)
The single-prolonged stress (SPS) model is a tool in which a complex stressor is consistently presented. This tool is used to explore the complexity of PTSD, particularly its impaired fear extinction.