Molecular scale electronics, also called single-molecule electronics, is a branch of nanotechnology that uses single molecules, or nanoscale collections of single molecules, as electronic components. Because single molecules constitute the smallest stable structures imaginable, this miniaturization is the ultimate goal for shrinking electrical circuits.
The field is often termed simply as "molecular electronics", but this term is also used to refer to the distantly related field of conductive polymers and organic electronics, which uses the properties of molecules to affect the bulk properties of a material. A nomenclature distinction has been suggested so that molecular materials for electronics refers to this latter field of bulk applications, while molecular scale electronics refers to the nanoscale single-molecule applications treated here.[1][2]
Fundamental concepts
Conventional electronics have traditionally been made from bulk materials. Ever since their invention in 1958, the performance and complexity of integrated circuits has undergone exponential growth, a trend named Moore’s law, as feature sizes of the embedded components have shrunk accordingly. As the structures shrink, the sensitivity to deviations increases. In a few technology generations, when the minimum feature sizes reaches 13 nm, the composition of the devices must be controlled to a precision of a few atoms[3] for the devices to work. With bulk methods growing increasingly demanding and costly as they near inherent limits, the idea was born that the components could instead be built up atom by atom in a chemistry lab (bottom up) versus carving them out of bulk material (top down). This is the idea behind molecular electronics, with the ultimate miniaturization being components contained in single molecules.In single-molecule electronics, the bulk material is replaced by single molecules. Instead of forming structures by removing or applying material after a pattern scaffold, the atoms are put together in a chemistry lab. In this way, billions of billions of copies are made simultaneously (typically more than 1020 molecules are made at once) while the composition of molecules are controlled down to the last atom. The molecules used have properties that resemble traditional electronic components such as a wire, transistor or rectifier.
Single-molecule electronics is an emerging field, and entire electronic circuits consisting exclusively of molecular sized compounds are still very far from being realized. However, the unceasing demand for more computing power, along with the inherent limits of lithographic methods as of 2016, make the transition seem unavoidable. Currently, the focus is on discovering molecules with interesting properties and on finding ways to obtain reliable and reproducible contacts between the molecular components and the bulk material of the electrodes.
Theoretical basis
Molecular electronics operates in the quantum realm of distances less than 100 nanometers. The miniaturization down to single molecules brings the scale down to a regime where quantum mechanics effects are important. In conventional electronic components, electrons can be filled in or drawn out more or less like a continuous flow of electric charge. In contrast, in molecular electronics the transfer of one electron alters the system significantly. For example, when an electron has been transferred from a source electrode to a molecule, the molecule gets charged up, which makes it far harder for the next electron to transfer (see also Coulomb blockade). The significant amount of energy due to charging must be accounted for when making calculations about the electronic properties of the setup, and is highly sensitive to distances to conducting surfaces nearby.The theory of single-molecule devices is especially interesting since the system under consideration is an open quantum system in nonequilibrium (driven by voltage). In the low bias voltage regime, the nonequilibrium nature of the molecular junction can be ignored, and the current-voltage traits of the device can be calculated using the equilibrium electronic structure of the system. However, in stronger bias regimes a more sophisticated treatment is required, as there is no longer a variational principle. In the elastic tunneling case (where the passing electron does not exchange energy with the system), the formalism of Rolf Landauer can be used to calculate the transmission through the system as a function of bias voltage, and hence the current. In inelastic tunneling, an elegant formalism based on the non-equilibrium Green's functions of Leo Kadanoff and Gordon Baym, and independently by Leonid Keldysh was advanced by Ned Wingreen and Yigal Meir. This Meir-Wingreen formulation has been used to great success in the molecular electronics community to examine the more difficult and interesting cases where the transient electron exchanges energy with the molecular system (for example through electron-phonon coupling or electronic excitations).
Further, connecting single molecules reliably to a larger scale circuit has proven a great challenge, and constitutes a significant hindrance to commercialization.
Examples
Common for molecules used in molecular electronics is that the structures contain many alternating double and single bonds (see also Conjugated system). This is done because such patterns delocalize the molecular orbitals, making it possible for electrons to move freely over the conjugated area.Wires
This animation of a rotating carbon nanotube shows its 3D structure.
The sole purpose of molecular wires is to electrically connect different parts of a molecular electrical circuit. As the assembly of these and their connection to a macroscopic circuit is still not mastered, the focus of research in single-molecule electronics is primarily on the functionalized molecules: molecular wires are characterized by containing no functional groups and are hence composed of plain repetitions of a conjugated building block. Among these are the carbon nanotubes that are quite large compared to the other suggestions but have shown very promising electrical properties.
The main problem with the molecular wires is to obtain good electrical contact with the electrodes so that electrons can move freely in and out of the wire.
Transistors
Single-molecule transistors are fundamentally different from the ones known from bulk electronics. The gate in a conventional (field-effect) transistor determines the conductance between the source and drain electrode by controlling the density of charge carriers between them, whereas the gate in a single-molecule transistor controls the possibility of a single electron to jump on and off the molecule by modifying the energy of the molecular orbitals. One of the effects of this difference is that the single-molecule transistor is almost binary: it is either on or off. This opposes its bulk counterparts, which have quadratic responses to gate voltage.It is the quantization of charge into electrons that is responsible for the markedly different behavior compared to bulk electronics. Because of the size of a single molecule, the charging due to a single electron is significant and provides means to turn a transistor on or off (see Coulomb blockade). For this to work, the electronic orbitals on the transistor molecule cannot be too well integrated with the orbitals on the electrodes. If they are, an electron cannot be said to be located on the molecule or the electrodes and the molecule will function as a wire.
A popular group of molecules, that can work as the semiconducting channel material in a molecular transistor, is the oligopolyphenylenevinylenes (OPVs) that works by the Coulomb blockade mechanism when placed between the source and drain electrode in an appropriate way.[4] Fullerenes work by the same mechanism and have also been commonly used.
Semiconducting carbon nanotubes have also been demonstrated to work as channel material but although molecular, these molecules are sufficiently large to behave almost as bulk semiconductors.
The size of the molecules, and the low temperature of the measurements being conducted, makes the quantum mechanical states well defined. Thus, it is being researched if the quantum mechanical properties can be used for more advanced purposes than simple transistors (e.g. spintronics).
Physicists at the University of Arizona, in collaboration with chemists from the University of Madrid, have designed a single-molecule transistor using a ring-shaped molecule similar to benzene. Physicists at Canada's National Institute for Nanotechnology have designed a single-molecule transistor using styrene. Both groups expect (the designs were experimentally unverified as of June 2005) their respective devices to function at room temperature, and to be controlled by a single electron.[5]
Rectifiers (diodes)
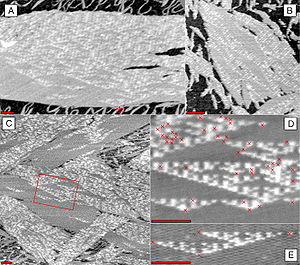
Hydrogen can be removed from individual tetraphenylporphyrin (H2TPP) molecules by applying excess voltage to the tip of a scanning tunneling microscope (STAM, a); this removal alters the current-voltage (I-V) curves of TPP molecules, measured using the same STM tip, from diode-like (red curve in b) to resistor-like (green curve). Image (c) shows a row of TPP, H2TPP and TPP molecules. While scanning image (d), excess voltage was applied to H2TPP
at the black dot, which instantly removed hydrogen, as shown in the
bottom part of (d) and in the re-scan image (e). Such manipulations can
be used in single-molecule electronics.[6]
Molecular rectifiers are mimics of their bulk counterparts and have an asymmetric construction so that the molecule can accept electrons in one end but not the other. The molecules have an electron donor (D) in one end and an electron acceptor (A) in the other. This way, the unstable state D+ – A− will be more readily made than D− – A+. The result is that an electric current can be drawn through the molecule if the electrons are added through the acceptor end, but less easily if the reverse is attempted.
Methods
One of the biggest problems with measuring on single molecules is to establish reproducible electrical contact with only one molecule and doing so without shortcutting the electrodes. Because the current photolithographic technology is unable to produce electrode gaps small enough to contact both ends of the molecules tested (on the order of nanometers), alternative strategies are applied.Molecular gaps
One way to produce electrodes with a molecular sized gap between them is break junctions, in which a thin electrode is stretched until it breaks. Another is electromigration. Here a current is led through a thin wire until it melts and the atoms migrate to produce the gap. Further, the reach of conventional photolithography can be enhanced by chemically etching or depositing metal on the electrodes.Probably the easiest way to conduct measurements on several molecules is to use the tip of a scanning tunneling microscope (STM) to contact molecules adhered at the other end to a metal substrate.[7]
Anchoring
A popular way to anchor molecules to the electrodes is to make use of sulfur's high chemical affinity to gold. In these setups, the molecules are synthesized so that sulfur atoms are placed strategically to function as crocodile clips connecting the molecules to the gold electrodes. Though useful, the anchoring is non-specific and thus anchors the molecules randomly to all gold surfaces. Further, the contact resistance is highly dependent on the precise atomic geometry around the site of anchoring and thereby inherently compromises the reproducibility of the connection.To circumvent the latter issue, experiments has shown that fullerenes could be a good candidate for use instead of sulfur because of the large conjugated π-system that can electrically contact many more atoms at once than one atom of sulfur.[8]
Fullerene nanoelectronics
In polymers, classical organic molecules are composed of both carbon and hydrogen (and sometimes additional compounds such as nitrogen, chlorine or sulphur). They are obtained from petrol and can often be synthesized in large amounts. Most of these molecules are insulating when their length exceeds a few nanometers. However, naturally occurring carbon is conducting, especially graphite recovered from coal or encountered otherwise. From a theoretical viewpoint, graphite is a semi-metal, a category in between metals and semi-conductors. It has a layered structure, each sheet being one atom thick. Between each sheet, the interactions are weak enough to allow an easy manual cleavage.Tailoring the graphite sheet to obtain well defined nanometer-sized objects remains a challenge. However, by the close of the twentieth century, chemists were exploring methods to fabricate extremely small graphitic objects that could be considered single molecules. After studying the interstellar conditions under which carbon is known to form clusters, Richard Smalley's group (Rice University, Texas) set up an experiment in which graphite was vaporized via laser irradiation. Mass spectrometry revealed that clusters containing specific magic numbers of atoms were stable, especially those clusters of 60 atoms. Harry Kroto, an English chemist who assisted in the experiment, suggested a possible geometry for these clusters – atoms covalently bound with the exact symmetry of a soccer ball. Coined buckminsterfullerenes, buckyballs, or C60, the clusters retained some properties of graphite, such as conductivity. These objects were rapidly envisioned as possible building blocks for molecular electronics.
Problems
Artifacts
When trying to measure electronic traits of molecules, artificial phenomena can occur that can be hard to distinguish from truly molecular behavior.[9] Before they were discovered, these artifacts have mistakenly been published as being features pertaining to the molecules in question.Applying a voltage drop on the order of volts across a nanometer sized junction results in a very strong electrical field. The field can cause metal atoms to migrate and eventually close the gap by a thin filament, which can be broken again when carrying a current. The two levels of conductance imitate molecular switching between a conductive and an isolating state of a molecule.
Another encountered artifact is when the electrodes undergo chemical reactions due to the high field strength in the gap. When the voltage bias is reversed, the reaction will cause hysteresis in the measurements that can be interpreted as being of molecular origin.
A metallic grain between the electrodes can act as a single electron transistor by the mechanism described above, thus resembling the traits of a molecular transistor. This artifact is especially common with nanogaps produced by the electromigration method.
Commercialization
One of the biggest hindrances for single-molecule electronics to be commercially exploited is the lack of methods to connect a molecular sized circuit to bulk electrodes in a way that gives reproducible results. At the current state, the difficulty of connecting single molecules vastly outweighs any possible performance increase that could be gained from such shrinkage. The difficulties grow worse if the molecules are to have a certain spatial orientation and/or have multiple poles to connect.Also problematic is that some measurements on single molecules are carried out in cryogenic temperatures (near absolute zero), which is very energy consuming. This is done to reduce signal noise enough to measure the faint currents of single molecules.
History and recent progress
Graphical representation of a rotaxane, useful as a molecular switch.
In their treatment of so-called donor-acceptor complexes in the 1940s, Robert Mulliken and Albert Szent-Györgyi advanced the concept of charge transfer in molecules. They subsequently further refined the study of both charge transfer and energy transfer in molecules. Likewise, a 1974 paper from Mark Ratner and Ari Aviram illustrated a theoretical molecular rectifier.[10] In 1988, Aviram described in detail a theoretical single-molecule field-effect transistor. Further concepts were proposed by Forrest Carter of the Naval Research Laboratory, including single-molecule logic gates. A wide range of ideas were presented, under his aegis, at a conference entitled Molecular Electronic Devices in 1988.[11] These were all theoretical constructs and not concrete devices. The direct measurement of the electronic traits of individual molecules awaited the development of methods for making molecular-scale electrical contacts. This was no easy task. Thus, the first experiment directly-measuring the conductance of a single molecule was only reported in 1995 on a single C60 molecule by C. Joachim and J. K. Gimzewsky in their seminal Physical Revie Letter paper and later in 1997 by Mark Reed and co-workers on a few hundred molecules. Since then, this branch of the field has advanced rapidly. Likewise, as it has grown possible to measure such properties directly, the theoretical predictions of the early workers have been confirmed substantially.
Recent progress in nanotechnology and nanoscience has facilitated both experimental and theoretical study of molecular electronics. Development of the scanning tunneling microscope (STM) and later the atomic force microscope (AFM) have greatly facilitated manipulating single-molecule electronics. Also, theoretical advances in molecular electronics have facilitated further understanding of non-adiabatic charge transfer events at electrode-electrolyte interfaces.[12][13]
The concept of molecular electronics was first published in 1974 when Aviram and Ratner suggested an organic molecule that could work as a rectifier.[14] Having both huge commercial and fundamental interest, much effort was put into proving its feasibility, and 16 years later in 1990, the first demonstration of an intrinsic molecular rectifier was realized by Ashwell and coworkers for a thin film of molecules.
The first measurement of the conductance of a single molecule was realised in 1994 by C. Joachim and J. K. Gimzewski and published in 1995 (see the corresponding Phys. Rev. Lett. paper). This was the conclusion of 10 years of research started at IBM TJ Watson, using the scanning tunnelling microscope tip apex to switch a single molecule as already explored by A. Aviram, C. Joachim and M. Pomerantz at the end of the 80's (see their seminal Chem. Phys. Lett. paper during this period). The trick was to use an UHV Scanning Tunneling microscope to allow the tip apex to gently touch the top of a single C
60 molecule adsorbed on an Au(110) surface. A resistance of 55 MOhms was recorded along with a low voltage linear I-V. The contact was certified by recording the I-z current distance property, which allows measurement of the deformation of the C
60 cage under contact. This first experiment was followed by the reported result using a mechanical break junction method to connect two gold electrodes to a sulfur-terminated molecular wire by Mark Reed and James Tour in 1997.[15]
A single-molecule amplifier was implemented by C. Joachim and J.K. Gimzewski in IBM Zurich. This experiment, involving one C
60 molecule, demonstrated that one such molecule can provide gain in a circuit via intramolecular quantum interference effects alone.
A collaboration of researchers at Hewlett-Packard (HP) and University of California, Los Angeles (UCLA), led by James Heath, Fraser Stoddart, R. Stanley Williams, and Philip Kuekes, has developed molecular electronics based on rotaxanes and catenanes.
Work is also occurring on the use of single-wall carbon nanotubes as field-effect transistors. Most of this work is being done by International Business Machines (IBM).
Some specific reports of a field-effect transistor based on molecular self-assembled monolayers were shown to be fraudulent in 2002 as part of the Schön scandal.[16]
Until recently entirely theoretical, the Aviram-Ratner model for a unimolecular rectifier has been confirmed unambiguously in experiments by a group led by Geoffrey J. Ashwell at Bangor University, UK.[17][18][19] Many rectifying molecules have so far been identified, and the number and efficiency of these systems is growing rapidly.
Supramolecular electronics is a new field involving electronics at a supramolecular level.
An important issue in molecular electronics is the determination of the resistance of a single molecule (both theoretical and experimental). For example, Bumm, et al. used STM to analyze a single molecular switch in a self-assembled monolayer to determine how conductive such a molecule can be.[20] Another problem faced by this field is the difficulty of performing direct characterization since imaging at the molecular scale is often difficult in many experimental devices.