The history of genetics dates from the classical era with contributions by Hippocrates, Aristotle and Epicurus. Modern biology began with the work of the Augustinian friar Gregor Johann Mendel. His work on pea plants, published in 1866,what is now Mendelian inheritance. Some theories of heredity suggest in the centuries before and for several decades after Mendel's work.
The year 1900 marked the "rediscovery of Mendel" by Hugo de Vries, Carl Correns and Erich von Tschermak, and by 1915 the basic principles of Mendelian genetics had been applied to a wide variety of organisms—most notably the fruit fly Drosophila melanogaster. Led by Thomas Hunt Morgan and his fellow "drosophilists", geneticists developed the Mendelian model, which was widely accepted by 1925. Alongside experimental work, mathematicians developed the statistical framework of population genetics, bringing genetic explanations into the study of evolution.
With the basic patterns of genetic inheritance established, many biologists turned to investigations of the physical nature of the gene. In the 1940s and early 1950s, experiments pointed to DNA as the portion of chromosomes (and perhaps other nucleoproteins) that held genes. A focus on new model organisms such as viruses and bacteria, along with the discovery of the double helical structure of DNA in 1953, marked the transition to the era of molecular genetics.
In the following years, chemists developed techniques for sequencing both nucleic acids and proteins, while others worked out the relationship between the two forms of biological molecules: the genetic code. The regulation of gene expression became a central issue in the 1960s; by the 1970s gene expression could be controlled and manipulated through genetic engineering. In the last decades of the 20th century, many biologists focused on large-scale genetics projects, sequencing entire genomes.
The year 1900 marked the "rediscovery of Mendel" by Hugo de Vries, Carl Correns and Erich von Tschermak, and by 1915 the basic principles of Mendelian genetics had been applied to a wide variety of organisms—most notably the fruit fly Drosophila melanogaster. Led by Thomas Hunt Morgan and his fellow "drosophilists", geneticists developed the Mendelian model, which was widely accepted by 1925. Alongside experimental work, mathematicians developed the statistical framework of population genetics, bringing genetic explanations into the study of evolution.
With the basic patterns of genetic inheritance established, many biologists turned to investigations of the physical nature of the gene. In the 1940s and early 1950s, experiments pointed to DNA as the portion of chromosomes (and perhaps other nucleoproteins) that held genes. A focus on new model organisms such as viruses and bacteria, along with the discovery of the double helical structure of DNA in 1953, marked the transition to the era of molecular genetics.
In the following years, chemists developed techniques for sequencing both nucleic acids and proteins, while others worked out the relationship between the two forms of biological molecules: the genetic code. The regulation of gene expression became a central issue in the 1960s; by the 1970s gene expression could be controlled and manipulated through genetic engineering. In the last decades of the 20th century, many biologists focused on large-scale genetics projects, sequencing entire genomes.
Pre-Mendelian ideas on heredity
Ancient theories
Aristotle's model of transmission of movements from parents to child, and of form from the father. The model is not fully symmetric.
In the Charaka Samhita of 300CE, ancient Indian medical writers saw the characteristics of the child as determined by four factors: 1) those from the mother’s reproductive material, (2) those from the father’s sperm, (3) those from the diet of the pregnant mother and (4) those accompanying the soul which enters into the foetus. Each of these four factors had four parts creating sixteen factors of which the karma of the parents and the soul determined which attributes predominated and thereby gave the child its characteristics.
In the 9th century CE, the Afro-Arab writer Al-Jahiz considered the effects of the environment on the likelihood of an animal to survive. In 1000 CE, the Arab physician, Abu al-Qasim al-Zahrawi (known as Albucasis in the West) was the first physician to describe clearly the hereditary nature of haemophilia in his Al-Tasrif. In 1140 CE, Judah HaLevi described dominant and recessive genetic traits in The Kuzari.
Plant systematics and hybridization
In the 18th century, with increased knowledge of plant and animal diversity and the accompanying increased focus on taxonomy, new ideas about heredity began to appear. Linnaeus and others (among them Joseph Gottlieb Kölreuter, Carl Friedrich von Gärtner, and Charles Naudin) conducted extensive experiments with hybridization, especially species hybrids. Species hybridizers described a wide variety of inheritance phenomena, include hybrid sterility and the high variability of back-crosses.Plant breeders were also developing an array of stable varieties in many important plant species. In the early 19th century, Augustin Sageret established the concept of dominance, recognizing that when some plant varieties are crossed, certain characteristics (present in one parent) usually appear in the offspring; he also found that some ancestral characteristics found in neither parent may appear in offspring. However, plant breeders made little attempt to establish a theoretical foundation for their work or to share their knowledge with current work of physiology, although Gartons Agricultural Plant Breeders in England explained their system.
Mendel
Blending inheritance leads to the averaging out of every characteristic, which would make evolution by natural selection impossible.
In breeding experiments between 1856 and 1865, Gregor Mendel first traced inheritance patterns of certain traits in pea plants and showed that they obeyed simple statistical rules with some traits being dominant and others being recessive. These patterns of Mendelian inheritance demonstrated that application of statistics to inheritance could be highly useful; they also contradicted 19th century theories of blending inheritance as the traits remained discrete through multiple generation of hybridization.
From his statistical analysis Mendel defined a concept that he described as a character (which in his mind holds also for "determinant of that character"). In only one sentence of his historical paper he used the term "factors" to designate the "material creating" the character: " So far as experience goes, we find it in every case confirmed that constant progeny can only be formed when the egg cells and the fertilizing pollen are of like character, so that both are provided with the material for creating quite similar individuals, as is the case with the normal fertilization of pure species. We must therefore regard it as certain that exactly similar factors must be at work also in the production of the constant forms in the hybrid plants."(Mendel, 1866).
Mendel's work was published in 1866 as "Versuche über Pflanzen-Hybriden" (Experiments on Plant Hybridization) in the Verhandlungen des Naturforschenden Vereins zu Brünn (Proceedings of the Natural History Society of Brünn), following two lectures he gave on the work in early 1866.
Post-Mendel, pre-rediscovery
Pangenesis
Diagram of Charles Darwin's pangenesis theory. Every part of the body emits tiny particles, gemmules, which migrate to the gonads
and contribute to the fertilised egg and so to the next generation. The
theory implied that changes to the body during an organism's life would
be inherited, as proposed in Lamarckism.
Mendel's work was published in a relatively obscure scientific journal, and it was not given any attention in the scientific community. Instead, discussions about modes of heredity were galvanized by Darwin's theory of evolution by natural selection, in which mechanisms of non-Lamarckian heredity seemed to be required. Darwin's own theory of heredity, pangenesis, did not meet with any large degree of acceptance. A more mathematical version of pangenesis, one which dropped much of Darwin's Lamarckian holdovers, was developed as the "biometrical" school of heredity by Darwin's cousin, Francis Galton.
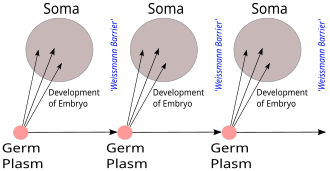
Germ plasm
August Weismann's germ plasm theory. The hereditary material, the germ plasm, is confined to the gonads. Somatic cells (of the body) develop afresh in each generation from the germ plasm.
In 1883 August Weismann conducted experiments involving breeding mice whose tails had been surgically removed. His results — that surgically removing a mouse's tail had no effect on the tail of its offspring — challenged the theories of pangenesis and Lamarckism, which held that changes to an organism during its lifetime could be inherited by its descendants. Weismann proposed the germ plasm theory of inheritance, which held that hereditary information was carried only in sperm and egg cells.
Rediscovery of Mendel
Hugo de Vries wondered what the nature of germ plasm might be, and in particular he wondered whether or not germ plasm was mixed like paint or whether the information was carried in discrete packets that remained unbroken. In the 1890s he was conducting breeding experiments with a variety of plant species and in 1897 he published a paper on his results that stated that each inherited trait was governed by two discrete particles of information, one from each parent, and that these particles were passed along intact to the next generation. In 1900 he was preparing another paper on his further results when he was shown a copy of Mendel's 1866 paper by a friend who thought it might be relevant to de Vries's work. He went ahead and published his 1900 paper without mentioning Mendel's priority. Later that same year another botanist, Carl Correns, who had been conducting hybridization experiments with maize and peas, was searching the literature for related experiments prior to publishing his own results when he came across Mendel's paper, which had results similar to his own. Correns accused de Vries of appropriating terminology from Mendel's paper without crediting him or recognizing his priority. At the same time another botanist, Erich von Tschermak was experimenting with pea breeding and producing results like Mendel's. He too discovered Mendel's paper while searching the literature for relevant work. In a subsequent paper de Vries praised Mendel and acknowledged that he had only extended his earlier work.Emergence of molecular genetics
After the rediscovery of Mendel's work there was a feud between William Bateson and Pearson over the hereditary mechanism, solved by Ronald Fisher in his work "The Correlation Between Relatives on the Supposition of Mendelian Inheritance".
Thomas Hunt Morgan discovered sex linked inheritance of the white eyed mutation in the fruit fly Drosophila in 1910, implying the gene was on the sex chromosome.
In 1910, Thomas Hunt Morgan showed that genes reside on specific chromosomes. He later showed that genes occupy specific locations on the chromosome. With this knowledge, Morgan and his students began the first chromosomal map of the fruit fly Drosophila melanogaster. In 1928, Frederick Griffith showed that genes could be transferred. In what is now known as Griffith's experiment, injections into a mouse of a deadly strain of bacteria that had been heat-killed transferred genetic information to a safe strain of the same bacteria, killing the mouse.
A series of subsequent discoveries led to the realization decades later that the genetic material is made of DNA (deoxyribonucleic acid). In 1941, George Wells Beadle and Edward Lawrie Tatum showed that mutations in genes caused errors in specific steps in metabolic pathways. This showed that specific genes code for specific proteins, leading to the "one gene, one enzyme" hypothesis. Oswald Avery, Colin Munro MacLeod, and Maclyn McCarty showed in 1944 that DNA holds the gene's information. In 1952, Rosalind Franklin and Raymond Gosling produced a strikingly clear x-ray diffraction pattern indicating a helical form, and in 1953, James D. Watson and Francis Crick demonstrated the molecular structure of DNA. Together, these discoveries established the central dogma of molecular biology, which states that proteins are translated from RNA which is transcribed by DNA. This dogma has since been shown to have exceptions, such as reverse transcription in retroviruses.
In 1972, Walter Fiers and his team at the University of Ghent were the first to determine the sequence of a gene: the gene for bacteriophage MS2 coat protein. Richard J. Roberts and Phillip Sharp discovered in 1977 that genes can be split into segments. This led to the idea that one gene can make several proteins. The successful sequencing of many organisms' genomes has complicated the molecular definition of the gene. In particular, genes do not always sit side by side on DNA like discrete beads. Instead, regions of the DNA producing distinct proteins may overlap, so that the idea emerges that "genes are one long continuum". It was first hypothesized in 1986 by Walter Gilbert that neither DNA nor protein would be required in such a primitive system as that of a very early stage of the earth if RNA could serve both as a catalyst and as genetic information storage processor.
The modern study of genetics at the level of DNA is known as molecular genetics and the synthesis of molecular genetics with traditional Darwinian evolution is known as the modern evolutionary synthesis.
Early timeline
- 1856-1863: Mendel studied the inheritance of traits between generations based on experiments involving garden pea plants. He deduced that there is a certain tangible essence that is passed on between generations from both parents. Mendel established the basic principles of inheritance, namely, the principles of dominance, independent assortment, and segregation.
- 1866: Austrian Augustinian monk Gregor Mendel's paper, Experiments on Plant Hybridization, published.
- 1869: Friedrich Miescher discovers a weak acid in the nuclei of white blood cells that today we call DNA. In 1871 he isolated cell nuclei, separated the nucleic cells from bandages and then treated them with pepsin (an enzyme which breaks down proteins). From this, he recovered an acidic substance which he called "nuclein."
- 1880-1890: Walther Flemming, Eduard Strasburger, and Edouard Van Beneden elucidate chromosome distribution during cell division.
- 1889: Richard Altmann purified protein free DNA. However, the nucleic acid was not as pure as he had assumed. It was determined later to contain a large amount of protein.
- 1889: Hugo de Vries postulates that "inheritance of specific traits in organisms comes in particles", naming such particles "(pan)genes".
- 1902: Archibald Garrod discovered inborn errors of metabolism. An explanation for epistasis is an important manifestation of Garrod’s research, albeit indirectly. When Garrod studied alkaptonuria, a disorder that makes urine quickly turn black due to the presence of gentesate, he noticed that it was prevalent among populations whose parents were closely related.
- 1903: Walter Sutton and Theodor Boveri independently hypothesizes that chromosomes, which segregate in a Mendelian fashion, are hereditary units. Boveri was studying sea urchins when he found that all the chromosomes in the sea urchins had to be present for proper embryonic development to take place. Sutton's work with grasshoppers showed that chromosomes occur in matched pairs of maternal and paternal chromosomes which separate during meiosis. He concluded that this could be "the physical basis of the Mendelian law of heredity."
- 1905: William Bateson coins the term "genetics" in a letter to Adam Sedgwick (Zoologist) and at a meeting in 1906.
- 1908: G.H. Hardy and Wilhelm Weinberg proposed the Hardy-Weinberg equilibrium model which describes the frequencies of alleles in the gene pool of a population, which are under certain specific conditions, as constant and at a state of equilibrium from generation to generation unless specific disturbing influences are introduced.
- 1910: Thomas Hunt Morgan shows that genes reside on chromosomes while determining the nature of sex-linked traits by studying Drosophila melanogaster. He determined that the white-eyed mutant was sex-linked based on Mendelian's principles of segregation and independent assortment.
- 1911: Alfred Sturtevant, one of Morgan's students, invented the procedure of linkage mapping which is based on the frequency of recombination.
- 1913: Alfred Sturtevant makes the first genetic map of a chromosome.
- 1913: Gene maps show chromosomes containing linear arranged genes.
- 1918: Ronald Fisher publishes "The Correlation Between Relatives on the Supposition of Mendelian Inheritance" the modern synthesis of genetics and evolutionary biology starts. See population genetics.
- 1920: Lysenkoism Started, during Lysenkoism they stated that the hereditary factor are not only in the nucleus, but also in the cytoplasm, though they called it living protoplasm.
- 1923: Frederick Griffith studied bacterial transformation and observed that DNA carries genes responsible for pathogenicity.
In Griffith's experiment,
mice are injected with dead bacteria of one strain and live bacteria of
another, and develop an infection of the dead strain's type.
- 1928: Frederick Griffith discovers that hereditary material from dead bacteria can be incorporated into live bacteria.
- 1930s–1950s: Joachim Hämmerling conducted experiments with Acetabularia in which he began to distinguish the contributions of the nucleus and the cytoplasm substances (later discovered to be DNA and mRNA, respectively) to cell morphogenesis and development.
- 1931: Crossing over is identified as the cause of recombination; the first cytological demonstration of this crossing over was performed by Barbara McClintock and Harriet Creighton.
- 1933: Jean Brachet, while studying virgin sea urchin eggs, suggested that DNA is found in cell nucleus and that RNA is present exclusively in the cytoplasm. At the time, "yeast nucleic acid" (RNA) was thought to occur only in plants, while "thymus nucleic acid" (DNA) only in animals. The latter was thought to be a tetramer, with the function of buffering cellular pH.
- 1933: Thomas Morgan received the Nobel prize for linkage mapping. His work elucidated the role played by the chromosome in heredity.
- 1941: Edward Lawrie Tatum and George Wells Beadle show that genes code for proteins.
- 1943: Luria–Delbrück experiment: this experiment showed that genetic mutations conferring resistance to bacteriophage arise in the absence of selection, rather than being a response to selection.
The DNA era
- 1944: The Avery–MacLeod–McCarty experiment isolates DNA as the genetic material (at that time called transforming principle).
- 1947: Salvador Luria discovers reactivation of irradiated phage, stimulating numerous further studies of DNA repair processes in bacteriophage, and other organisms, including humans.
- 1948: Barbara McClintock discovers transposons in maize.
- 1950: Erwin Chargaff determined the pairing method of nitrogenous bases. Chargaff and his team studied the DNA from multiple organisms and found three things (also known as Chargaff's rules). First, the concentration of the pyrimidines (guanine and adenine) are always found in the same amount as one another. Second, the concentration of purines (cytosine and thymidine) are also always the same. Lastly, Chargaff and his team found the proportion of pyrimidines and purines correspond each other.
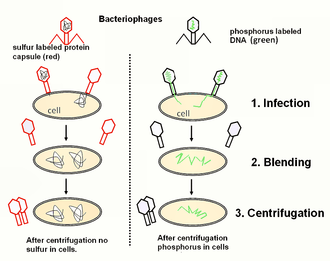
- 1952: The Hershey–Chase experiment proves the genetic information of phages (and, by implication, all other organisms) to be DNA.
- 1952: an X-ray diffraction image of DNA taken by Raymond Gosling in May 1952, a student supervised by Rosalind Franklin
- 1953: DNA structure is resolved to be a double helix by Rosalind Franklin, James Watson and Francis Crick.
- 1955: Alexander R. Todd determined the chemical makeup of nitrogenous bases. Todd also successfully synthesized adenosine triphosphate (ATP) and flavin adenine dinucleotide (FAD). He was awarded the Nobel prize in Chemistry in 1957 for his contributions in the scientific knowledge of nucleotides and nucleotide co-enzymes.
- 1955: Joe Hin Tjio, while working in Albert Levan's lab, determined the number of chromosomes in humans to be of 46. Tjio was attempting to refine an established technique to separate chromosomes onto glass slides by conducting a study of human embryonic lung tissue, when he saw that there were 46 chromosomes rather than 48. This revolutionized the world of cytogenetics.
- 1957: Arthur Kornberg with Severo Ochoa synthesized DNA in a test tube after discovering the means by which DNA is duplicated . DNA polymerase 1 established requirements for in vitro synthesis of DNA. Kornberg and Ochoa were awarded the Nobel Prize in 1959 for this work.
- 1957/1958: Robert W. Holley, Marshall Nirenberg, Har Gobind Khorana proposed the nucleotide sequence of the tRNA molecule. Francis Crick had proposed the requirement of some kind of adapter molecule and it was soon identified by Holey, Nirenberg and Khorana. These scientists help explain the link between a messenger RNA nucleotide sequence and a polypeptide sequence. In the experiment, they purified tRNAs from yeast cells and were awarded the Nobel prize in 1968.
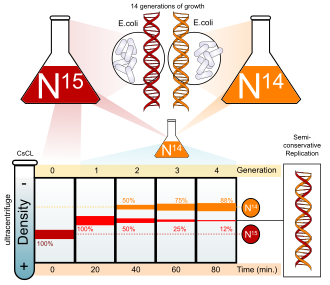
Meselson–Stahl experiment demonstrates DNA is semiconservatively replicated.
- 1958: The Meselson–Stahl experiment demonstrates that DNA is semiconservatively replicated.
- 1960: Jacob and collaborators discover the operon, a group of genes whose expression is coordinated by an operator.
- 1961: Francis Crick and Sydney Brenner discovered frame shift mutations. In the experiment, proflavin-induced mutations of the T4 bacteriophage gene (rIIB) were isolated. Proflavin causes mutations by inserting itself between DNA bases, typically resulting in insertion or deletion of a single base pair. The mutants could not produce functional rIIB protein. These mutations were used to demonstrate that three sequential bases of the rIIB gene’s DNA specify each successive amino acid of the encoded protein. Thus the genetic code is a triplet code, where each triplet (called a codon) specifies a particular amino acid.
- 1961: Sydney Brenner, Francois Jacob and Matthew Meselson identified the function of messenger RNA.
- 1961 - 1967: Combined efforts of scientists "crack" the genetic code, including Marshall Nirenberg, Har Gobind Khorana, Sydney Brenner & Francis Crick.
- 1964: Howard Temin showed using RNA viruses that the direction of DNA to RNA transcription can be reversed.
- 1964: Lysenkoism Ended.
- 1966: Marshall W. Nirenberg, Philip Leder, Har Gobind Khorana cracked the genetic code by using RNA homopolymer and heteropolymer experiments, through which they figured out which triplets of RNA were translated into what amino acids in yeast cells.
- 1969: Molecular hybridization of radioactive DNA to the DNA of cytological preparation. by Pardue, M. L. and Gall, J. G.
- 1970: Restriction enzymes were discovered in studies of a bacterium, Haemophilus influenzae, by Hamilton O. Smith and Daniel Nathans, enabling scientists to cut and paste DNA.
- 1972: Stanley Norman Cohen and Herbert Boyer at UCSF and Stanford University constructed Recombinant DNA which can be formed by using restriction Endonuclease to cleave the DNA and DNA ligase to reattach the "sticky ends" into a bacterial plasmid.
The genomics era
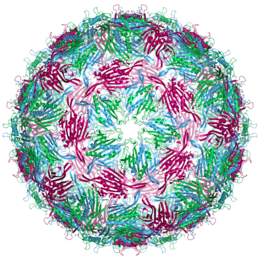
In 1972, the first gene was sequenced: the gene for bacteriophage MS2 coat protein (3 chains in different colours).
- 1972: Walter Fiers and his team were the first to determine the sequence of a gene: the gene for bacteriophage MS2 coat protein.
- 1976: Walter Fiers and his team determine the complete nucleotide-sequence of bacteriophage MS2-RNA.
- 1976: Yeast genes expressed in E. coli for the first time.
- 1977: DNA is sequenced for the first time by Fred Sanger, Walter Gilbert, and Allan Maxam working independently. Sanger's lab sequence the entire genome of bacteriophage Φ-X174.
- In the late 1970s: nonisotopic methods of nucleic acid labeling were developed. The subsequent improvements in the detection of reporter molecules using immunocytochemistry and immunofluorescence,in conjunction with advances in fluorescence microscopy and image analysis, have made the technique safer, faster and reliable.
- 1980: Paul Berg, Walter Gilbert and Frederick Sanger developed methods of mapping the structure of DNA. In 1972, recombinant DNA molecules were produced in Paul Berg’s Stanford University laboratory. Berg was awarded the 1980 Nobel Prize in Chemistry for constructing recombinant DNA molecules that contained phage lambda genes inserted into the small circular DNA mol.
- 1980: Stanley Norman Cohen and Herbert Boyer received first U.S. patent for gene cloning, by proving the successful outcome of cloning a plasmid and expressing a foreign gene in bacteria to produce a "protein foreign to a unicellular organism." These two scientist were able to replicate proteins such as HGH, Erythropoietin and Insulin. The patent earned about $300 million in licensing royalties for Stanford.
- 1982: The U.S. Food and Drug Administration (FDA) approved the release of the first genetically engineered human insulin, originally biosynthesized using recombination DNA methods by Genentech in 1978. Once approved, the cloning process lead to mass production of humulin (under license by Eli Lilly & Co.).
- 1983: Kary Banks Mullis invents the polymerase chain reaction enabling the easy amplification of DNA.
- 1983: Barbara McClintock was awarded the Nobel Prize in Physiology or Medicine for her discovery of mobile genetic elements. McClintock studied transposon-mediated mutation and chromosome breakage in maize and published her first report in 1948 on transposable elements or transposons. She found that transposons were widely observed in corn, although her ideas weren't widely granted attention until the 1960s and 1970s when the same phenomenon was discovered in bacteria and Drosophila melanogaster.
Display of VNTR allele lengths on a chromatogram, a technology used in DNA fingerprinting
- 1985: Alec Jeffreys announced DNA fingerprinting method. Jeffreys was studying DNA variation and the evolution of gene families in order to understand disease causing genes. In an attempt to develop a process to isolate many mini-satellites at once using chemical probes, Jeffreys took x-ray films of the DNA for examination and noticed that mini-satellite regions differ greatly from one person to another. In a DNA fingerprinting technique, a DNA sample is digested by treatment with specific nucleases or Restriction endonuclease and then the fragments are separated by electrophoresis producing a template distinct to each individual banding pattern of the gel.
- 1986: Jeremy Nathans found genes for color vision and color blindness, working with David Hogness, Douglas Vollrath and Ron Davis as they were studying the complexity of the retina.
- 1987: Yoshizumi Ishino accidentally discovers and describes part of a DNA sequence which later will be called CRISPR.
- 1989: Thomas Cech discovered that RNA can catalyze chemical reactions, making for one of the most important breakthroughs in molecular genetics, because it elucidates the true function of poorly understood segments of DNA.
- 1989: The human gene that encodes the CFTR protein was sequenced by Francis Collins and Lap-Chee Tsui. Defects in this gene cause cystic fibrosis.
- 1992: American and British scientists unveiled a technique for testing embryos in-vitro (Amniocentesis) for genetic abnormalities such as Cystic fibrosis and Hemophilia.
- 1993: Phillip Allen Sharp and Richard Roberts awarded the Nobel Prize for the discovery that genes in DNA are made up of introns and exons. According to their findings not all the nucleotides on the RNA strand (product of DNA transcription) are used in the translation process. The intervening sequences in the RNA strand are first spliced out so that only the RNA segment left behind after splicing would be translated to polypeptides.
- 1994: The first breast cancer gene is discovered. BRCA I, was discovered by researchers at the King laboratory at UC Berkeley in 1990 but was first cloned in 1994. BRCA II, the second key gene in the manifestation of breast cancer was discovered later in 1994 by Professor Michael Stratton and Dr. Richard Wooster.
- 1995: The genome of bacterium Haemophilus influenzae is the first genome of a free living organism to be sequenced.
- 1996: Saccharomyces cerevisiae , a yeast species, is the first eukaryote genome sequence to be released.
- 1996: Alexander Rich discovered the Z-DNA, a type of DNA which is in a transient state, that is in some cases associated with DNA transcription. The Z-DNA form is more likely to occur in regions of DNA rich in cytosine and guanine with high salt concentrations.
- 1997: Dolly the sheep was cloned by Ian Wilmut and colleagues from the Roslin Institute in Scotland.
- 1998: The first genome sequence for a multicellular eukaryote, Caenorhabditis elegans, is released.
- 2000: The full genome sequence of Drosophila melanogaster is completed.
- 2001: First draft sequences of the human genome are released simultaneously by the Human Genome Project and Celera Genomics.
- 2001: Francisco Mojica and Rudd Jansen propose the acronym CRISPR to describe a family of bacterial DNA sequences that can be used to specifically change genes within organisms.
Francis Collins announces the successful completion of the Human Genome Project in 2003
- 2003 (14 April): Successful completion of Human Genome Project with 99% of the genome sequenced to a 99.99% accuracy.
- 2004: Merck introduced a vaccine for Human Papillomavirus which promised to protect women against infection with HPV 16 and 18, which inactivates tumor suppressor genes and together cause 70% of cervical cancers.
- 2007: Michael Worobey traced the evolutionary origins of HIV by analyzing its genetic mutations, which revealed that HIV infections had occurred in the United States as early as the 1960s.
- 2007: Timothy Ray Brown becomes the first person cured from HIV/AIDS through a Hematopoietic stem cell transplantation.
- 2008: Houston-based Introgen developed Advexin (FDA Approval pending), the first gene therapy for cancer and Li-Fraumeni syndrome, utilizing a form of Adenovirus to carry a replacement gene coding for the p53 protein.
- 2010: transcription activator-like effector nucleases (or TALENs) are first used to cut specific sequences of DNA.
- 2016: A genome is sequenced in outer space for the first time, with NASA astronaut Kate Rubins using a MinION device aboard the International Space Station.[85]