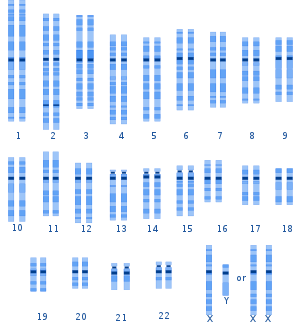A graphical representation of the typical human karyotype.
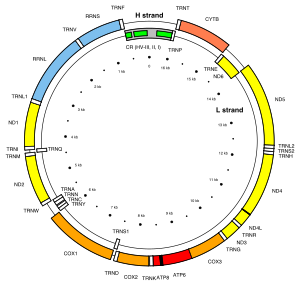
The human mitochondrial DNA.
Human genetic variation is the genetic differences in and among populations. There may be multiple variants of any given gene in the human population (alleles), a situation called polymorphism.
No two humans are genetically identical. Even monozygotic twins (who develop from one zygote) have infrequent genetic differences due to mutations occurring during development and gene copy-number variation. Differences between individuals, even closely related individuals, are the key to techniques such as genetic fingerprinting.
As of 2017, there are a total of 324 million known variants from sequenced human genomes.[2]
As of 2015, the typical difference between the genomes of two
individuals was estimated at 20 million base pairs (or 0.6% of the total
of 3.2 billion base pairs).
Alleles occur at different frequencies in different human populations. Populations that are more geographically and ancestrally remote
tend to differ more. The differences between populations represent a
small proportion of overall human genetic variation. Populations also
differ in the quantity of variation among their members.
The greatest divergence between populations is found in sub-Saharan Africa, consistent with the recent African origin of non-African populations.
Populations also vary in the proportion and locus of introgressed genes they received by archaic admixture both inside and outside of Africa.
The study of human genetic variation has evolutionary
significance and medical applications. It can help scientists understand
ancient human population migrations as well as how human groups are
biologically related to one another. For medicine, study of human
genetic variation may be important because some disease-causing alleles
occur more often in people from specific geographic regions. New
findings show that each human has on average 60 new mutations compared
to their parents.
Causes of variation
Causes of differences between individuals include independent assortment, the exchange of genes (crossing over and recombination) during reproduction (through meiosis) and various mutational events.
There are at least three reasons why genetic variation exists between populations. Natural selection
may confer an adaptive advantage to individuals in a specific
environment if an allele provides a competitive advantage. Alleles under
selection are likely to occur only in those geographic regions where
they confer an advantage. A second important process is genetic drift, which is the effect of random changes in the gene pool, under conditions where most mutations are neutral
(that is, they do not appear to have any positive or negative selective
effect on the organism). Finally, small migrant populations have
statistical differences called the founder effect—from
the overall populations where they originated; when these migrants
settle new areas, their descendant population typically differs from
their population of origin: different genes predominate and it is less
genetically diverse.
In humans, the main cause is genetic drift. Serial founder effects
and past small population size (increasing the likelihood of genetic
drift) may have had an important influence in neutral differences
between populations. The second main cause of genetic variation is due to the high degree of neutrality of most mutations.
A small, but significant number of genes appear to have undergone
recent natural selection, and these selective pressures are sometimes
specific to one region.
Measures of variation
Genetic variation among humans occurs on many scales, from gross alterations in the human karyotype to single nucleotide changes. Chromosome abnormalities are detected in 1 of 160 live human births. Apart from sex chromosome disorders, most cases of aneuploidy result in death of the developing fetus (miscarriage); the most common extra autosomal chromosomes among live births are 21, 18 and 13.
Nucleotide diversity
is the average proportion of nucleotides that differ between two
individuals. As of 2004, the human nucleotide diversity was estimated to
be 0.1% to 0.4% of base pairs. In 2015, the 1000 Genomes Project,
which sequenced one thousand individuals from 26 human populations,
found that "a typical [individual] genome differs from the reference
human genome at 4.1 million to 5.0 million sites … affecting 20 million
bases of sequence"; the latter figure corresponds to 0.6% of total
number of base pairs.
Nearly all (>99.9%) of these sites are small differences, either
single nucleotide polymorphisms or brief insertions or deletions (indels) in the genetic sequence, but structural variations account for a greater number of base-pairs than the SNPs and indels.
As of 2017, the Single Nucleotide Polymorphism Database (dbSNP), which lists SNP and other variants, listed 324 million variants found in sequenced human genomes.
Single nucleotide polymorphisms
DNA molecule 1 differs from DNA molecule 2 at a single base-pair location (a C/T polymorphism).
A single nucleotide polymorphism
(SNP) is a difference in a single nucleotide between members of one
species that occurs in at least 1% of the population. The 2,504
individuals characterized by the 1000 Genomes Project had 84.7 million
SNPs among them. SNPs are the most common type of sequence variation, estimated in 1998 to account for 90% of all sequence variants. Other sequence variations are single base exchanges, deletions and insertions. SNPs occur on average about every 100 to 300 bases and so are the major source of heterogeneity.
A functional, or non-synonymous, SNP is one that affects some factor such as gene splicing or messenger RNA, and so causes a phenotypic difference between members of the species. About 3% to 5% of human SNPs are functional (see International HapMap Project). Neutral, or synonymous SNPs are still useful as genetic markers in genome-wide association studies, because of their sheer number and the stable inheritance over generations.
A coding SNP is one that occurs inside a gene. There are 105 Human Reference SNPs that result in premature stop codons
in 103 genes. This corresponds to 0.5% of coding SNPs. They occur due
to segmental duplication in the genome. These SNPs result in loss of
protein, yet all these SNP alleles are common and are not purified in negative selection.
Structural variation
Structural variation is the variation in structure of an organism's chromosome. Structural variations, such as copy-number variation and deletions, inversions, insertions and duplications,
account for much more human genetic variation than single nucleotide
diversity. This was concluded in 2007 from analysis of the diploid full sequences of the genomes of two humans: Craig Venter and James D. Watson. This added to the two haploid sequences which were amalgamations of sequences from many individuals, published by the Human Genome Project and Celera Genomics respectively.
According to the 1000 Genomes Project, a typical human has 2,100
to 2,500 structural variations, which include approximately 1,000 large
deletions, 160 copy-number variants, 915 Alu insertions, 128 L1 insertions, 51 SVA insertions, 4 NUMTs, and 10 inversions.
Copy number variation
A copy-number variation (CNV) is a difference in the genome due to
deleting or duplicating large regions of DNA on some chromosome. It is
estimated that 0.4% of the genomes of unrelated humans differ with
respect to copy number. When copy number variation is included,
human-to-human genetic variation is estimated to be at least 0.5% (99.5%
similarity). Copy number variations are inherited but can also arise during development.
A visual map with the regions with high genomic variation of the modern-human reference assembly relatively to a
Neanderthal of 50k has been built by Pratas et al.
Epigenetics
Epigenetic variation is variation in the chemical tags that attach to DNA and affect how genes get read. The tags, "called epigenetic markings, act as switches that control how genes can be read." At some alleles, the epigenetic state of the DNA, and associated phenotype, can be inherited across generations of individuals.
Genetic variability
Genetic variability is a measure of the tendency of individual genotypes in a population to vary (become different) from one another. Variability is different from genetic diversity,
which is the amount of variation seen in a particular population. The
variability of a trait is how much that trait tends to vary in response
to environmental and genetic influences.
Clines
In biology, a cline is a continuum of species, populations, races,
varieties, or forms of organisms that exhibit gradual phenotypic and/or
genetic differences over a geographical area, typically as a result of
environmental heterogeneity.
In the scientific study of human genetic variation, a gene cline can be
rigorously defined and subjected to quantitative metrics.
Haplogroups
In the study of molecular evolution, a haplogroup is a group of similar haplotypes that share a common ancestor with a single nucleotide polymorphism (SNP) mutation. Haplogroups pertain to deep ancestral origins dating back thousands of years.
The most commonly studied human haplogroups are Y-chromosome (Y-DNA) haplogroups and mitochondrial DNA (mtDNA) haplogroups, both of which can be used to define genetic populations. Y-DNA is passed solely along the patrilineal line, from father to son, while mtDNA is passed down the matrilineal line, from mother to both daughter or son. The Y-DNA and mtDNA may change by chance mutation at each generation.
Variable number tandem repeats
A variable number tandem repeat (VNTR) is the variation of length of a tandem repeat. A tandem repeat is the adjacent repetition of a short nucleotide sequence. Tandem repeats exist on many chromosomes, and their length varies between individuals. Each variant acts as an inherited allele, so they are used for personal or parental identification. Their analysis is useful in genetics and biology research, forensics, and DNA fingerprinting.
Short tandem repeats (about 5 base pairs) are called microsatellites, while longer ones are called minisatellites.
History and geographic distribution
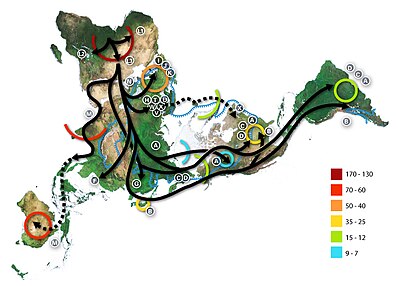
Map of the migration of modern humans out of Africa, based on mitochondrial DNA. Colored rings indicate thousand years before present.
Genetic distance map by Magalhães et al. (2012)
Recent African origin of modern humans
The recent African origin of modern humans paradigm assumes the dispersal of non-African populations of anatomically modern humans
after 70,000 years ago. Dispersal within Africa occurred significantly
earlier, at least 130,000 years ago. The "out of Africa" theory
originates in the 19th century, as a tentative suggestion in Charles
Darwin's Descent of Man,
but remained speculative until the 1980s when it was supported by study
of present-day mitochondrial DNA, combined with evidence from physical anthropology of archaic specimens.
According to a 2000 study of Y-chromosome sequence variation,
human Y-chromosomes trace ancestry to Africa, and the descendants of
the derived lineage left Africa and eventually were replaced by archaic
human Y-chromosomes in Eurasia. The study also shows that a minority of
contemporary populations in East Africa and the Khoisan
are the descendants of the most ancestral patrilineages of anatomically
modern humans that left Africa 35,000 to 89,000 years ago.
Other evidence supporting the theory is that variations in skull
measurements decrease with distance from Africa at the same rate as the
decrease in genetic diversity. Human genetic diversity decreases in
native populations with migratory distance from Africa, and this is
thought to be due to bottlenecks during human migration, which are events that temporarily reduce population size.
A 2009 genetic clustering
study, which genotyped 1327 polymorphic markers in various African
populations, identified six ancestral clusters. The clustering
corresponded closely with ethnicity, culture and language. A 2018 whole genome sequencing
study of the world's populations observed similar clusters among the
populations in Africa. At K=9, distinct ancestral components defined the
Afroasiatic-speaking populations inhabiting North Africa and Northeast Africa; the Nilo-Saharan-speaking populations in Northeast Africa and East Africa; the Ari populations in Northeast Africa; the Niger-Congo-speaking populations in West-Central Africa, West Africa, East Africa and Southern Africa; the Pygmy populations in Central Africa; and the Khoisan populations in Southern Africa.
Population genetics
Because of the common ancestry of all humans, only a small number of
variants have large differences in frequency between populations.
However, some rare variants in the world's human population are much
more frequent in at least one population (more than 5%).
Genetic variation
It is commonly assumed that early humans left Africa, and thus must
have passed through a population bottleneck before their
African-Eurasian divergence around 100,000 years ago (ca. 3,000
generations). The rapid expansion of a previously small population has two important effects on the distribution of genetic variation. First, the so-called founder effect
occurs when founder populations bring only a subset of the genetic
variation from their ancestral population. Second, as founders become
more geographically separated, the probability that two individuals from
different founder populations will mate becomes smaller. The effect of
this assortative mating is to reduce gene flow between geographical groups and to increase the genetic distance between groups.
The expansion of humans from Africa affected the distribution of
genetic variation in two other ways. First, smaller (founder)
populations experience greater genetic drift
because of increased fluctuations in neutral polymorphisms. Second, new
polymorphisms that arose in one group were less likely to be
transmitted to other groups as gene flow was restricted.
Populations in Africa tend to have lower amounts of linkage disequilibrium
than do populations outside Africa, partly because of the larger size
of human populations in Africa over the course of human history and
partly because the number of modern humans who left Africa to colonize
the rest of the world appears to have been relatively low.
In contrast, populations that have undergone dramatic size reductions
or rapid expansions in the past and populations formed by the mixture of
previously separate ancestral groups can have unusually high levels of
linkage disequilibrium
Distribution of variation
Human genetic variation calculated from genetic data representing 346 microsatellite
loci taken from 1484 individuals in 78 human populations. The upper
graph illustrates that as populations are further from East Africa, they
have declining genetic diversity as measured in average number of
microsatellite repeats at each of the loci. The bottom chart illustrates
isolation by distance.
Populations with a greater distance between them are more dissimilar
(as measured by the Fst statistic) than those which are geographically
close to one another. The horizontal axis of both charts is geographic
distance as measured along likely routes of human migration. (Chart from
Kanitz et al. 2018)
The distribution of genetic variants within and among human
populations are impossible to describe succinctly because of the
difficulty of defining a "population," the clinal nature of variation,
and heterogeneity across the genome (Long and Kittles 2003). In general,
however, an average of 85% of genetic variation exists within local
populations, ~7% is between local populations within the same continent,
and ~8% of variation occurs between large groups living on different
continents. The recent African origin
theory for humans would predict that in Africa there exists a great
deal more diversity than elsewhere and that diversity should decrease
the further from Africa a population is sampled.
Phenotypic variation
Sub-Saharan Africa has the most human genetic diversity and the same has been shown to hold true for phenotypic variation in skull form. Phenotype is connected to genotype through gene expression.
Genetic diversity decreases smoothly with migratory distance from that
region, which many scientists believe to be the origin of modern humans,
and that decrease is mirrored by a decrease in phenotypic variation.
Skull measurements are an example of a physical attribute whose
within-population variation decreases with distance from Africa.
The distribution of many physical traits resembles the distribution of genetic variation within and between human populations (American Association of Physical Anthropologists
1996; Keita and Kittles 1997). For example, ~90% of the variation in
human head shapes occurs within continental groups, and ~10% separates
groups, with a greater variability of head shape among individuals with
recent African ancestors (Relethford 2002).
A prominent exception to the common distribution of physical characteristics within and among groups is skin color.
Approximately 10% of the variance in skin color occurs within groups,
and ~90% occurs between groups (Relethford 2002). This distribution of
skin color and its geographic patterning — with people whose ancestors
lived predominantly near the equator having darker skin than those with
ancestors who lived predominantly in higher latitudes — indicate that
this attribute has been under strong selective pressure. Darker skin appears to be strongly selected for in equatorial regions to prevent sunburn, skin cancer, the photolysis of folate, and damage to sweat glands.
Understanding how genetic diversity in the human population
impacts various levels of gene expression is an active area of research.
While earlier studies focused on the relationship between DNA variation
and RNA expression, more recent efforts are characterizing the genetic
control of various aspects of gene expression including chromatin
states, translation, and protein levels.
A study published in 2007 found that 25% of genes showed different
levels of gene expression between populations of European and Asian
descent.
The primary cause of this difference in gene expression was thought to
be SNPs in gene regulatory regions of DNA. Another study published in
2007 found that approximately 83% of genes were expressed at different
levels among individuals and about 17% between populations of European
and African descent.
Wright's Fixation index as measure of variation
The population geneticist Sewall Wright developed the fixation index (often abbreviated to FST)
as a way of measuring genetic differences between populations. This
statistic is often used in taxonomy to compare differences between any
two given populations by measuring the genetic differences among and
between populations for individual genes, or for many genes
simultaneously.
It is often stated that the fixation index for humans is about 0.15.
This translates to an estimated 85% of the variation measured in the
overall human population is found within individuals of the same
population, and about 15% of the variation occurs between populations.
These estimates imply that any two individuals from different
populations are almost as likely to be more similar to each other than
either is to a member of their own group.
"The shared evolutionary history of living humans has resulted in a high relatedness among all living people, as indicated for example by the very low fixation index (FST) among living human populations."
Wright himself believed that values >0.25 represent very great genetic variation and that an FST
of 0.15–0.25 represented great variation. However, about 5% of human
variation occurs between populations within continents, therefore FST
values between continental groups of humans (or races) of as low as 0.1
(or possibly lower) have been found in some studies, suggesting more
moderate levels of genetic variation. Graves (1996) has countered that FST
should not be used as a marker of subspecies status, as the statistic
is used to measure the degree of differentiation between populations, although see also Wright (1978).
Jeffrey Long and Rick Kittles give a long critique of the application of FST
to human populations in their 2003 paper "Human Genetic Diversity and
the Nonexistence of Biological Races". They find that the figure of 85%
is misleading because it implies that all human populations contain on
average 85% of all genetic diversity. They argue the underlying
statistical model incorrectly assumes equal and independent histories of
variation for each large human population. A more realistic approach is
to understand that some human groups are parental to other groups and
that these groups represent paraphyletic groups to their descent groups. For example, under the recent African origin
theory the human population in Africa is paraphyletic to all other
human groups because it represents the ancestral group from which all
non-African populations derive, but more than that, non-African groups
only derive from a small non-representative sample of this African
population. This means that all non-African groups are more closely
related to each other and to some African groups (probably east
Africans) than they are to others, and further that the migration out of
Africa represented a genetic bottleneck,
with much of the diversity that existed in Africa not being carried out
of Africa by the emigrating groups. Under this scenario, human
populations do not have equal amounts of local variability, but rather
diminished amounts of diversity the further from Africa any population
lives. Long and Kittles find that rather than 85% of human genetic
diversity existing in all human populations, about 100% of human
diversity exists in a single African population, whereas only about 70%
of human genetic diversity exists in a population derived from New
Guinea. Long and Kittles argued that this still produces a global human
population that is genetically homogeneous compared to other mammalian
populations.
Archaic admixture
There is a hypothesis that anatomically modern humans interbred with Neanderthals during the Middle Paleolithic. In May 2010, the Neanderthal Genome Project presented genetic evidence that interbreeding did likely take place and that a small but significant
portion of Neanderthal admixture is present in the DNA of modern
Eurasians and Oceanians, and nearly absent in sub-Saharan African
populations.
Between 4% and 6% of the genome of Melanesians (represented by the Papua New Guinean and Bougainville Islander) are thought to derive from Denisova hominins
– a previously unknown species which shares a common origin with
Neanderthals. It was possibly introduced during the early migration of
the ancestors of Melanesians into Southeast Asia. This history of
interaction suggests that Denisovans once ranged widely over eastern
Asia.
Thus, Melanesians emerge as the most archaic-admixed population, having Denisovan/Neanderthal-related admixture of ~8%.
In a study published in 2013, Jeffrey Wall from University of
California studied whole sequence-genome data and found higher rates of
introgression in Asians compared to Europeans.
Hammer et al. tested the hypothesis that contemporary African genomes
have signatures of gene flow with archaic human ancestors and found
evidence of archaic admixture in African genomes, suggesting that modest
amounts of gene flow were widespread throughout time and space during
the evolution of anatomically modern humans.
Categorization of the world population
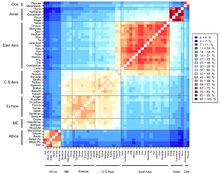
Chart showing human genetic clustering.
New data on human genetic variation has reignited the debate about a
possible biological basis for categorization of humans into races. Most
of the controversy surrounds the question of how to interpret the
genetic data and whether conclusions based on it are sound. Some
researchers argue that self-identified race can be used as an indicator
of geographic ancestry for certain health risks and medications.
Although the genetic differences among human groups are relatively small, these differences in certain genes such as duffy, ABCC11, SLC24A5, called ancestry-informative markers
(AIMs) nevertheless can be used to reliably situate many individuals
within broad, geographically based groupings. For example, computer
analyses of hundreds of polymorphic loci sampled in globally distributed
populations have revealed the existence of genetic clustering that
roughly is associated with groups that historically have occupied large
continental and subcontinental regions (Rosenberg et al. 2002; Bamshad et al. 2003).
Some commentators have argued that these patterns of variation
provide a biological justification for the use of traditional racial
categories. They argue that the continental clusterings correspond
roughly with the division of human beings into sub-Saharan Africans; Europeans, Western Asians, Central Asians, Southern Asians and Northern Africans; Eastern Asians, Southeast Asians, Polynesians and Native Americans; and other inhabitants of Oceania (Melanesians, Micronesians & Australian Aborigines) (Risch et al.
2002). Other observers disagree, saying that the same data undercut
traditional notions of racial groups (King and Motulsky 2002; Calafell
2003; Tishkoff and Kidd 2004).
They point out, for example, that major populations considered races or
subgroups within races do not necessarily form their own clusters.
Furthermore, because human genetic variation is clinal, many
individuals affiliate with two or more continental groups. Thus, the
genetically based "biogeographical ancestry" assigned to any given
person generally will be broadly distributed and will be accompanied by
sizable uncertainties (Pfaff et al. 2004).
In many parts of the world, groups have mixed in such a way that
many individuals have relatively recent ancestors from widely separated
regions. Although genetic analyses of large numbers of loci can produce
estimates of the percentage of a person's ancestors coming from various
continental populations (Shriver et al. 2003; Bamshad et al.
2004), these estimates may assume a false distinctiveness of the
parental populations, since human groups have exchanged mates from local
to continental scales throughout history (Cavalli-Sforza et al.
1994; Hoerder 2002). Even with large numbers of markers, information for
estimating admixture proportions of individuals or groups is limited,
and estimates typically will have wide confidence intervals (Pfaff et al. 2004).
Genetic clustering
Genetic data can be used to infer population structure and assign
individuals to groups that often correspond with their self-identified
geographical ancestry. Jorde and Wooding (2004) argued that
"Analysis of many loci now yields reasonably accurate estimates of
genetic similarity among individuals, rather than populations.
Clustering of individuals is correlated with geographic origin or
ancestry."
However, identification by geographic origin may quickly break down
when considering historical ancestry shared between individuals back in
time.
An analysis of autosomal SNP data from the International HapMap Project (Phase II) and CEPH Human Genome Diversity Panel samples was published in 2009.
The study of 53 populations taken from the HapMap and CEPH data (1138 unrelated individuals) suggested that natural selection
may shape the human genome much more slowly than previously thought,
with factors such as migration within and among continents more heavily
influencing the distribution of genetic variations.
A similar study published in 2010 found strong genome-wide evidence for
selection due to changes in ecoregion, diet, and subsistence
particularly in connection with polar ecoregions, with foraging, and
with a diet rich in roots and tubers. In a 2016 study, principal component analysis
of genome-wide data was capable of recovering previously-known targets
for positive selection (without prior definition of populations) as well
as a number of new candidate genes.
Forensic anthropology
Forensic anthropologists
can determine aspects of geographic ancestry (i.e. Asian, African, or
European) from skeletal remains with a high degree of accuracy by
analyzing skeletal measurements. According to some studies, individual test methods such as mid-facial measurements and femur
traits can identify the geographic ancestry and by extension the racial
category to which an individual would have been assigned during their
lifetime, with over 80% accuracy, and in combination can be even more
accurate. However, the skeletons of people who have recent ancestry in
different geographical regions can exhibit characteristics of more than
one ancestral group and, hence, cannot be identified as belonging to any
single ancestral group.
Triangle
plot shows average admixture of five North American ethnic groups.
Individuals that self-identify with each group can be found at many
locations on the map, but on average groups tend to cluster differently.
Gene flow and admixture
Gene flow between two populations reduces the average genetic
distance between the populations, only totally isolated human
populations experience no gene flow and most populations have continuous
gene flow with other neighboring populations which create the clinal
distribution observed for moth genetic variation. When gene flow takes
place between well-differentiated genetic populations the result is
referred to as "genetic admixture".
Admixture mapping is a technique used to study how genetic variants cause differences in disease rates between population.
Recent admixture populations that trace their ancestry to multiple
continents are well suited for identifying genes for traits and diseases
that differ in prevalence between parental populations.
African-American populations have been the focus of numerous population
genetic and admixture mapping studies, including studies of complex
genetic traits such as white cell count, body-mass index, prostate
cancer and renal disease.
An analysis of phenotypic and genetic variation including skin
color and socio-economic status was carried out in the population of
Cape Verde which has a well documented history of contact between
Europeans and Africans. The studies showed that pattern of admixture in
this population has been sex-biased and there is a significant
interactions between socio economic status and skin color independent of
the skin color and ancestry.
Another study shows an increased risk of graft-versus-host disease
complications after transplantation due to genetic variants in human
leukocyte antigen (HLA) and non-HLA proteins.
Health
Differences in allele frequencies contribute to group differences in the incidence of some monogenic diseases, and they may contribute to differences in the incidence of some common diseases.
For the monogenic diseases, the frequency of causative alleles usually
correlates best with ancestry, whether familial (for example, Ellis-van Creveld syndrome among the Pennsylvania Amish), ethnic (Tay–Sachs disease among Ashkenazi Jewish
populations), or geographical (hemoglobinopathies among people with
ancestors who lived in malarial regions). To the extent that ancestry
corresponds with racial or ethnic groups or subgroups, the incidence of
monogenic diseases can differ between groups categorized by race or
ethnicity, and health-care professionals typically take these patterns
into account in making diagnoses.
Even with common diseases involving numerous genetic variants and
environmental factors, investigators point to evidence suggesting the
involvement of differentially distributed alleles with small to moderate
effects. Frequently cited examples include hypertension (Douglas et al. 1996), diabetes (Gower et al. 2003), obesity (Fernandez et al. 2003), and prostate cancer (Platz et al.
2000). However, in none of these cases has allelic variation in a
susceptibility gene been shown to account for a significant fraction of
the difference in disease prevalence among groups, and the role of
genetic factors in generating these differences remains uncertain
(Mountain and Risch 2004).
Some other variations on the other hand are beneficial to human,
as they prevent certain diseases and increase the chance to adapt to the
environment. For example, mutation in CCR5 gene that protects against AIDS. CCR5 gene is absent on the surface of cell due to mutation. Without CCR5 gene on the surface, there is nothing for HIV
viruses to grab on and bind into. Therefore the mutation on CCR5 gene
decreases the chance of an individual’s risk with AIDS. The mutation in
CCR5 is also quite popular in certain areas, with more than 14% of the
population carry the mutation in Europe and about 6–10% in Asia and North Africa.
HIV attachment
Apart from mutations, many genes that may have aided humans in
ancient times plague humans today. For example, it is suspected that
genes that allow humans to more efficiently process food are those that
make people susceptible to obesity and diabetes today.
Neil Risch of Stanford University
has proposed that self-identified race/ethnic group could be a valid
means of categorization in the USA for public health and policy
considerations. A 2002 paper by Noah Rosenberg's
group makes a similar claim: "The structure of human populations is
relevant in various epidemiological contexts. As a result of variation
in frequencies of both genetic and nongenetic risk factors, rates of
disease and of such phenotypes as adverse drug response vary across
populations. Further, information about a patient’s population of origin
might provide health care practitioners with information about risk
when direct causes of disease are unknown."
Genome projects
Human genome projects are scientific endeavors that determine or study the structure of the human genome. The Human Genome Project was a landmark genome project.