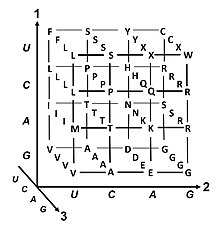
The genetic code is the set of rules used by living cells to translate information encoded within genetic material (DNA or RNA sequences of nucleotide triplets, or codons) into proteins. Translation is accomplished by the ribosome, which links proteinogenic amino acids in an order specified by messenger RNA (mRNA), using transfer RNA (tRNA) molecules to carry amino acids and to read the mRNA three nucleotides at a time. The genetic code is highly similar among all organisms and can be expressed in a simple table with 64 entries.
The codons specify which amino acid will be added next during protein biosynthesis. With some exceptions, a three-nucleotide codon in a nucleic acid sequence specifies a single amino acid. The vast majority of genes are encoded with a single scheme (see the RNA codon table). That scheme is often referred to as the canonical or standard genetic code, or simply the genetic code, though variant codes (such as in mitochondria) exist.
History
Efforts to understand how proteins are encoded began after DNA's structure was discovered in 1953. The key discoverers, English biophysicist Francis Crick and American biologist James Watson, working together at the Cavendish Laboratory of the University of Cambridge, hypothesied that information flows from DNA and that there is a link between DNA and proteins. Soviet-American physicist George Gamow was the first to give a workable scheme for protein synthesis from DNA. He postulated that sets of three bases (triplets) must be employed to encode the 20 standard amino acids used by living cells to build proteins, which would allow a maximum of 43 = 64 amino acids. He named this DNA–protein interaction (the original genetic code) as the "diamond code."
In 1954, Gamow created an informal scientific organisation the RNA Tie Club, as suggested by Watson, for scientists of different persuasions who were interested in how proteins were synthesised from genes. However, the club could have only 20 permanent members to represent each of the 20 amino acids; and four additional honorary members to represent the four nucleotides of DNA.
The first scientific contribution of the club, later recorded as "one of the most important unpublished articles in the history of science" and "the most famous unpublished paper in the annals of molecular biology," was made by Crick. Crick presented a type-written paper titled "On Degenerate Templates and the Adaptor Hypothesis: A Note for the RNA Tie Club" to the members of the club in January 1955, which "totally change the way we thought about protein synthesis", as Watson recalled. The hypothesis states that the triplet code was not passed on to amino acids as Gamow thought, but carried by a different molecule, an adaptor, that interacts with amino acids. The adaptor was later identified as tRNA.
Codons
The Crick, Brenner, Barnett and Watts-Tobin experiment first demonstrated that codons consist of three DNA bases. Marshall Nirenberg and Heinrich J. Matthaei were the first to reveal the nature of a codon in 1961.
They used a cell-free system to translate a poly-uracil RNA sequence (i.e., UUUUU...) and discovered that the polypeptide that they had synthesized consisted of only the amino acid phenylalanine. They thereby deduced that the codon UUU specified the amino acid phenylalanine.
This was followed by experiments in Severo Ochoa's laboratory that demonstrated that the poly-adenine RNA sequence (AAAAA...) coded for the polypeptide poly-lysine and that the poly-cytosine RNA sequence (CCCCC...) coded for the polypeptide poly-proline. Therefore, the codon AAA specified the amino acid lysine, and the codon CCC specified the amino acid proline. Using various copolymers most of the remaining codons were then determined.
Subsequent work by Har Gobind Khorana identified the rest of the genetic code. Shortly thereafter, Robert W. Holley determined the structure of transfer RNA (tRNA), the adapter molecule that facilitates the process of translating RNA into protein. This work was based upon Ochoa's earlier studies, yielding the latter the Nobel Prize in Physiology or Medicine in 1959 for work on the enzymology of RNA synthesis.
Extending this work, Nirenberg and Philip Leder revealed the code's triplet nature and deciphered its codons. In these experiments, various combinations of mRNA were passed through a filter that contained ribosomes, the components of cells that translate RNA into protein. Unique triplets promoted the binding of specific tRNAs to the ribosome. Leder and Nirenberg were able to determine the sequences of 54 out of 64 codons in their experiments. Khorana, Holley and Nirenberg received the 1968 Nobel for their work.
The three stop codons were named by discoverers Richard Epstein and Charles Steinberg. "Amber" was named after their friend Harris Bernstein, whose last name means "amber" in German. The other two stop codons were named "ochre" and "opal" in order to keep the "color names" theme.
Expanded genetic codes (synthetic biology)
In a broad academic audience, the concept of the evolution of the genetic code from the original and ambiguous genetic code to a well-defined ("frozen") code with the repertoire of 20 (+2) canonical amino acids is widely accepted. However, there are different opinions, concepts, approaches and ideas, which is the best way to change it experimentally. Even models are proposed that predict "entry points" for synthetic amino acid invasion of the genetic code.
Since 2001, 40 non-natural amino acids have been added into proteins by creating a unique codon (recoding) and a corresponding transfer-RNA:aminoacyl – tRNA-synthetase pair to encode it with diverse physicochemical and biological properties in order to be used as a tool to exploring protein structure and function or to create novel or enhanced proteins.
H. Murakami and M. Sisido extended some codons to have four and five bases. Steven A. Benner constructed a functional 65th (in vivo) codon.
In 2015 N. Budisa, D. Söll and co-workers reported the full substitution of all 20,899 tryptophan residues (UGG codons) with unnatural thienopyrrole-alanine in the genetic code of the bacterium Escherichia coli.
In 2016 the first stable semisynthetic organism was created. It was a (single cell) bacterium with two synthetic bases (called X and Y). The bases survived cell division.
In 2017, researchers in South Korea reported that they had engineered a mouse with an extended genetic code that can produce proteins with unnatural amino acids.
In May 2019, researchers reported the creation of a new "Syn61" strain of the bacterium Escherichia coli. This strain has a fully synthetic genome that is refactored (all overlaps expanded), recoded (removing the use of three out of 64 codons completely), and further modified to remove the now unnecessary tRNAs and release factors. It is fully viable and grows 1.6× slower than its wild-type counterpart "MDS42".
Features
Reading frame
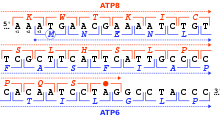
A reading frame is defined by the initial triplet of nucleotides from which translation starts. It sets the frame for a run of successive, non-overlapping codons, which is known as an "open reading frame" (ORF). For example, the string 5'-AAATGAACG-3' (see figure), if read from the first position, contains the codons AAA, TGA, and ACG ; if read from the second position, it contains the codons AAT and GAA ; and if read from the third position, it contains the codons ATG and AAC. Every sequence can, thus, be read in its 5' → 3' direction in three reading frames, each producing a possibly distinct amino acid sequence: in the given example, Lys (K)-Trp (W)-Thr (T), Asn (N)-Glu (E), or Met (M)-Asn (N), respectively (when translating with the vertebrate mitochondrial code). When DNA is double-stranded, six possible reading frames are defined, three in the forward orientation on one strand and three reverse on the opposite strand. Protein-coding frames are defined by a start codon, usually the first AUG (ATG) codon in the RNA (DNA) sequence.
In eukaryotes, ORFs in exons are often interrupted by introns.
Start and stop codons
Translation starts with a chain-initiation codon or start codon. The start codon alone is not sufficient to begin the process. Nearby sequences such as the Shine-Dalgarno sequence in E. coli and initiation factors are also required to start translation. The most common start codon is AUG, which is read as methionine or as formylmethionine (in bacteria, mitochondria, and plastids). Alternative start codons depending on the organism include "GUG" or "UUG"; these codons normally represent valine and leucine, respectively, but as start codons they are translated as methionine or formylmethionine.
The three stop codons have names: UAG is amber, UGA is opal (sometimes also called umber), and UAA is ochre. Stop codons are also called "termination" or "nonsense" codons. They signal release of the nascent polypeptide from the ribosome because no cognate tRNA has anticodons complementary to these stop signals, allowing a release factor to bind to the ribosome instead.
Effect of mutations
During the process of DNA replication, errors occasionally occur in the polymerization of the second strand. These errors, mutations, can affect an organism's phenotype, especially if they occur within the protein coding sequence of a gene. Error rates are typically 1 error in every 10–100 million bases—due to the "proofreading" ability of DNA polymerases.
Missense mutations and nonsense mutations are examples of point mutations that can cause genetic diseases such as sickle-cell disease and thalassemia respectively. Clinically important missense mutations generally change the properties of the coded amino acid residue among basic, acidic, polar or non-polar states, whereas nonsense mutations result in a stop codon.
Mutations that disrupt the reading frame sequence by indels (insertions or deletions) of a non-multiple of 3 nucleotide bases are known as frameshift mutations. These mutations usually result in a completely different translation from the original, and likely cause a stop codon to be read, which truncates the protein. These mutations may impair the protein's function and are thus rare in in vivo protein-coding sequences. One reason inheritance of frameshift mutations is rare is that, if the protein being translated is essential for growth under the selective pressures the organism faces, absence of a functional protein may cause death before the organism becomes viable. Frameshift mutations may result in severe genetic diseases such as Tay–Sachs disease.
Although most mutations that change protein sequences are harmful or neutral, some mutations have benefits. These mutations may enable the mutant organism to withstand particular environmental stresses better than wild type organisms, or reproduce more quickly. In these cases a mutation will tend to become more common in a population through natural selection. Viruses that use RNA as their genetic material have rapid mutation rates, which can be an advantage, since these viruses thereby evolve rapidly, and thus evade the immune system defensive responses. In large populations of asexually reproducing organisms, for example, E. coli, multiple beneficial mutations may co-occur. This phenomenon is called clonal interference and causes competition among the mutations.
Degeneracy
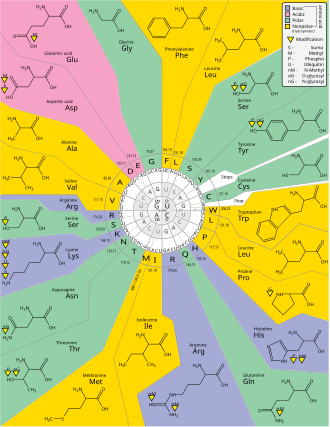
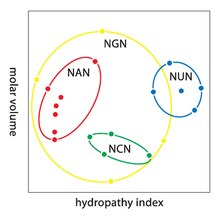
Degeneracy is the redundancy of the genetic code. This term was given by Bernfield and Nirenberg. The genetic code has redundancy but no ambiguity (see the codon tables below for the full correlation). For example, although codons GAA and GAG both specify glutamic acid (redundancy), neither specifies another amino acid (no ambiguity). The codons encoding one amino acid may differ in any of their three positions. For example, the amino acid leucine is specified by YUR or CUN (UUA, UUG, CUU, CUC, CUA, or CUG) codons (difference in the first or third position indicated using IUPAC notation), while the amino acid serine is specified by UCN or AGY (UCA, UCG, UCC, UCU, AGU, or AGC) codons (difference in the first, second, or third position). A practical consequence of redundancy is that errors in the third position of the triplet codon cause only a silent mutation or an error that would not affect the protein because the hydrophilicity or hydrophobicity is maintained by equivalent substitution of amino acids; for example, a codon of NUN (where N = any nucleotide) tends to code for hydrophobic amino acids. NCN yields amino acid residues that are small in size and moderate in hydropathicity; NAN encodes average size hydrophilic residues. The genetic code is so well-structured for hydropathicity that a mathematical analysis (Singular Value Decomposition) of 12 variables (4 nucleotides x 3 positions) yields a remarkable correlation (C = 0.95) for predicting the hydropathicity of the encoded amino acid directly from the triplet nucleotide sequence, without translation. Note in the table, below, eight amino acids are not affected at all by mutations at the third position of the codon, whereas in the figure above, a mutation at the second position is likely to cause a radical change in the physicochemical properties of the encoded amino acid. Nevertheless, changes in the first position of the codons are more important than changes in the second position on a global scale. The reason may be that charge reversal (from a positive to a negative charge or vice versa) can only occur upon mutations in the first position of certain codons, but not upon changes in the second position of any codon. Such charge reversal may have dramatic consequences for the structure or function of a protein. This aspect may have been largely underestimated by previous studies.
Codon usage bias
The frequency of codons, also known as codon usage bias, can vary from species to species with functional implications for the control of translation. The codon varies by organism; for example, most common proline codon in E. coli is CCG, whereas in humans this is the least used proline codon.
Human genome codon frequency table
|
|---|
Alternative genetic codes
Non-standard amino acids
In some proteins, non-standard amino acids are substituted for standard stop codons, depending on associated signal sequences in the messenger RNA. For example, UGA can code for selenocysteine and UAG can code for pyrrolysine. Selenocysteine came to be seen as the 21st amino acid, and pyrrolysine as the 22nd. Unlike selenocysteine, pyrrolysine-encoded UAG is translated with the participation of a dedicated aminoacyl-tRNA synthetase. Both selenocysteine and pyrrolysine may be present in the same organism. Although the genetic code is normally fixed in an organism, the achaeal prokaryote Acetohalobium arabaticum can expand its genetic code from 20 to 21 amino acids (by including pyrrolysine) under different conditions of growth.
Variations
There was originally a simple and widely accepted argument that the genetic code should be universal: namely, that any variation in the genetic code would be lethal to the organism (although Crick had stated that viruses were an exception). This is known as the "frozen accident" argument for the universality of the genetic code. However, in his seminal paper on the origins of the genetic code in 1968, Francis Crick still stated that the universality of the genetic code in all organisms was an unproven assumption, and was probably not true in some instances. He predicted that "The code is universal (the same in all organisms) or nearly so". The first variation was discovered in 1979, by researchers studying human mitochondrial genes. Many slight variants were discovered thereafter, including various alternative mitochondrial codes. These minor variants for example involve translation of the codon UGA as tryptophan in Mycoplasma species, and translation of CUG as a serine rather than leucine in yeasts of the "CTG clade" (such as Candida albicans). Because viruses must use the same genetic code as their hosts, modifications to the standard genetic code could interfere with viral protein synthesis or functioning. However, viruses such as totiviruses have adapted to the host's genetic code modification. In bacteria and archaea, GUG and UUG are common start codons. In rare cases, certain proteins may use alternative start codons. Surprisingly, variations in the interpretation of the genetic code exist also in human nuclear-encoded genes: In 2016, researchers studying the translation of malate dehydrogenase found that in about 4% of the mRNAs encoding this enzyme the stop codon is naturally used to encode the amino acids tryptophan and arginine. This type of recoding is induced by a high-readthrough stop codon context and it is referred to as functional translational readthrough.
Despite these differences, all known naturally occurring codes are very similar. The coding mechanism is the same for all organisms: three-base codons, tRNA, ribosomes, single direction reading and translating single codons into single amino acids. The most extreme variations occur in certain ciliates where the meaning of stop codons depends on their position within mRNA. When close to the 3' end they act as terminators while in internal positions they either code for amino acids as in Condylostoma magnum or trigger ribosomal frameshifting as in Euplotes.
The origins and variation of the genetic code, including the mechanisms behind the evolvability of the genetic code, have been widely studied, and some studies have been done experimentally evolving the genetic code of some organisms.
Inferrence
Variant genetic codes used by an organism can be inferred by identifying highly conserved genes encoded in that genome, and comparing its codon usage to the amino acids in homologous proteins of other organisms. For example, the program FACIL infers a genetic code by searching which amino acids in homologous protein domains are most often aligned to every codon. The resulting amino acid (or stop codon) probabilities for each codon are displayed in a genetic code logo.
As of January 2022, the most complete survey of genetic codes is done by Shulgina and Eddy, who screened 250,000 prokaryotic genomes using their Codetta tool. This tool uses a similar approach to FACIL with a larger Pfam database. Despite the NCBI already providing 33 translation tables, the authors were able to find new 5 genetic code variations (corroborated by tRNA mutations) and correct several misattributions.
Origin
The genetic code is a key part of the history of life, according to one version of which self-replicating RNA molecules preceded life as we know it. This is the RNA world hypothesis. Under this hypothesis, any model for the emergence of the genetic code is intimately related to a model of the transfer from ribozymes (RNA enzymes) to proteins as the principal enzymes in cells. In line with the RNA world hypothesis, transfer RNA molecules appear to have evolved before modern aminoacyl-tRNA synthetases, so the latter cannot be part of the explanation of its patterns.
A hypothetical randomly evolved genetic code further motivates a biochemical or evolutionary model for its origin. If amino acids were randomly assigned to triplet codons, there would be 1.5 × 1084 possible genetic codes. This number is found by calculating the number of ways that 21 items (20 amino acids plus one stop) can be placed in 64 bins, wherein each item is used at least once. However, the distribution of codon assignments in the genetic code is nonrandom. In particular, the genetic code clusters certain amino acid assignments.
Amino acids that share the same biosynthetic pathway tend to have the same first base in their codons. This could be an evolutionary relic of an early, simpler genetic code with fewer amino acids that later evolved to code a larger set of amino acids. It could also reflect steric and chemical properties that had another effect on the codon during its evolution. Amino acids with similar physical properties also tend to have similar codons, reducing the problems caused by point mutations and mistranslations.
Given the non-random genetic triplet coding scheme, a tenable hypothesis for the origin of genetic code could address multiple aspects of the codon table, such as absence of codons for D-amino acids, secondary codon patterns for some amino acids, confinement of synonymous positions to third position, the small set of only 20 amino acids (instead of a number approaching 64), and the relation of stop codon patterns to amino acid coding patterns.
Three main hypotheses address the origin of the genetic code. Many models belong to one of them or to a hybrid:
- Random freeze: the genetic code was randomly created. For example, early tRNA-like ribozymes may have had different affinities for amino acids, with codons emerging from another part of the ribozyme that exhibited random variability. Once enough peptides were coded for, any major random change in the genetic code would have been lethal; hence it became "frozen".
- Stereochemical affinity: the genetic code is a result of a high affinity between each amino acid and its codon or anti-codon; the latter option implies that pre-tRNA molecules matched their corresponding amino acids by this affinity. Later during evolution, this matching was gradually replaced with matching by aminoacyl-tRNA synthetases.
- Optimality: the genetic code continued to evolve after its initial creation, so that the current code maximizes some fitness function, usually some kind of error minimization.
Hypotheses have addressed a variety of scenarios:
- Chemical principles govern specific RNA interaction with amino acids. Experiments with aptamers showed that some amino acids have a selective chemical affinity for their codons. Experiments showed that of 8 amino acids tested, 6 show some RNA triplet-amino acid association.
- Biosynthetic expansion. The genetic code grew from a simpler earlier code through a process of "biosynthetic expansion". Primordial life "discovered" new amino acids (for example, as by-products of metabolism) and later incorporated some of these into the machinery of genetic coding. Although much circumstantial evidence has been found to suggest that fewer amino acid types were used in the past, precise and detailed hypotheses about which amino acids entered the code in what order are controversial. However, several studies have suggested that Gly, Ala, Asp, Val, Ser, Pro, Glu, Leu, Thr may belong to a group of early-addition amino acids, whereas Cys, Met, Tyr, Trp, His, Phe may belong to a group of later-addition amino acids.
- Natural selection has led to codon assignments of the genetic code that minimize the effects of mutations. A recent hypothesis suggests that the triplet code was derived from codes that used longer than triplet codons (such as quadruplet codons). Longer than triplet decoding would increase codon redundancy and would be more error resistant. This feature could allow accurate decoding absent complex translational machinery such as the ribosome, such as before cells began making ribosomes.
- Information channels: Information-theoretic approaches model the process of translating the genetic code into corresponding amino acids as an error-prone information channel. The inherent noise (that is, the error) in the channel poses the organism with a fundamental question: how can a genetic code be constructed to withstand noise while accurately and efficiently translating information? These "rate-distortion" models suggest that the genetic code originated as a result of the interplay of the three conflicting evolutionary forces: the needs for diverse amino acids, for error-tolerance and for minimal resource cost. The code emerges at a transition when the mapping of codons to amino acids becomes nonrandom. The code's emergence is governed by the topology defined by the probable errors and is related to the map coloring problem.
- Game theory: Models based on signaling games combine elements of game theory, natural selection and information channels. Such models have been used to suggest that the first polypeptides were likely short and had non-enzymatic function. Game theoretic models suggested that the organization of RNA strings into cells may have been necessary to prevent "deceptive" use of the genetic code, i.e. preventing the ancient equivalent of viruses from overwhelming the RNA world.
- Stop codons: Codons for translational stops are also an interesting aspect to the problem of the origin of the genetic code. As an example for addressing stop codon evolution, it has been suggested that the stop codons are such that they are most likely to terminate translation early in the case of a frame shift error. In contrast, some stereochemical molecular models explain the origin of stop codons as "unassignable".