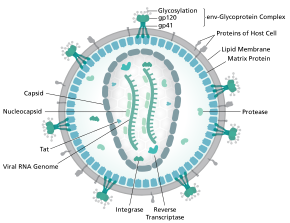
The genome and proteins of HIV (human immunodeficiency virus) have been the subject of extensive research since the discovery of the virus in 1983. "In the search for the causative agent, it was initially believed that the virus was a form of the Human T-cell leukemia virus (HTLV), which was known at the time to affect the human immune system and cause certain leukemias. However, researchers at the Pasteur Institute in Paris isolated a previously unknown and genetically distinct retrovirus in patients with AIDS which was later named HIV." Each virion comprises a viral envelope and associated matrix enclosing a capsid, which itself encloses two copies of the single-stranded RNA genome and several enzymes. The discovery of the virus itself occurred two years following the report of the first major cases of AIDS-associated illnesses.
Structure
The complete sequence of the HIV-1 genome, extracted from infectious virions, has been solved to single-nucleotide resolution. The HIV genome encodes a small number of viral proteins, invariably establishing cooperative associations among HIV proteins and between HIV and host proteins, to invade host cells and hijack their internal machineries. HIV is different in structure from other retroviruses. The HIV virion is ~100 nm in diameter. Its innermost region consists of a cone-shaped core that includes two copies of the (positive sense) ssRNA genome, the enzymes reverse transcriptase, integrase and protease, some minor proteins, and the major core protein. The genome of human immunodeficiency virus (HIV) encodes 8 viral proteins playing essential roles during the HIV life cycle.
HIV-1 is composed of two copies of noncovalently linked, unspliced, positive-sense single-stranded RNA enclosed by a conical capsid composed of the viral protein p24, typical of lentiviruses. The two copies of RNA strands are vital in contributing to HIV-1 recombination, which occurs during reverse transcription of viral replication. The containment of two copies of single-stranded RNA within a virion but the production of only a single DNA provirus is called pseudodiploidy. The RNA component is 9749 nucleotides long and bears a 5’ cap (Gppp), a 3’ poly(A) tail, and many open reading frames (ORFs). Viral structural proteins are encoded by long ORFs, whereas smaller ORFs encode regulators of the viral life cycle: attachment, membrane fusion, replication, and assembly.
The single-strand RNA is tightly bound to p7 nucleocapsid proteins, late assembly protein p6, and enzymes essential to the development of the virion, such as reverse transcriptase and integrase. Lysine tRNA is the primer of the magnesium-dependent reverse transcriptase. The nucleocapsid associates with the genomic RNA (one molecule per hexamer) and protects the RNA from digestion by nucleases. Also enclosed within the virion particle are Vif, Vpr, Nef, and viral protease. The envelope of the virion is formed by a plasma membrane of host cell origin, which is supported by a matrix composed of the viral p17 protein, ensuring the integrity of the virion particle. At the surface of the virion can be found a limited number of the envelope glycoprotein (Env) of HIV, a trimer formed by heterodimers of gp120 and gp41. Env is responsible for binding to its primary host receptor, CD4, and its co-receptor (mainly CCR5 or CXCR4), leading to viral entry into its target cell.
As the only proteins on the surface of the virus, the envelope glycoproteins (gp120 and gp41) are the major targets for HIV vaccine efforts. Over half of the mass of the trimeric envelope spike is N-linked glycans. The density is high as the glycans shield underlying viral protein from neutralisation by antibodies. This is one of the most densely glycosylated molecules known and the density is sufficiently high to prevent the normal maturation process of glycans during biogenesis in the endoplasmic reticulum and Golgi apparatus. The majority of the glycans are therefore stalled as immature 'high-mannose' glycans not normally present on secreted or cell surface human glycoproteins. The unusual processing and high density means that almost all broadly neutralising antibodies that have so far been identified (from a subset of patients that have been infected for many months to years) bind to or, are adapted to cope with, these envelope glycans.
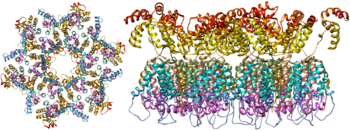
The molecular structure of the viral spike has now been determined by X-ray crystallography and cryo-electron microscopy. These advances in structural biology were made possible due to the development of stable recombinant forms of the viral spike by the introduction of an intersubunit disulphide bond and an isoleucine to proline mutation in gp41. The so-called SOSIP trimers not only reproduce the antigenic properties of the native viral spike but also display the same degree of immature glycans as presented on the native virus. Recombinant trimeric viral spikes are promising vaccine candidates as they display less non-neutralising epitopes than recombinant monomeric gp120 which act to suppress the immune response to target epitopes.
Genome organization
HIV has several major genes coding for structural proteins that are found in all retroviruses as well as several nonstructural ("accessory") genes unique to HIV. The HIV genome contains nine genes that encode fifteen viral proteins. These are synthesized as polyproteins which produce proteins for virion interior, called Gag, group specific antigen; the viral enzymes (Pol, polymerase) or the glycoproteins of the virion env (envelope). In addition to these, HIV encodes for proteins which have certain regulatory and auxiliary functions as well. HIV-1 has two important regulatory elements: Tat and Rev and few important accessory proteins such as Nef, Vpr, Vif and Vpu which are not essential for replication in certain tissues. The gag gene provides the basic physical infrastructure of the virus, and pol provides the basic mechanism by which retroviruses reproduce, while the others help HIV to enter the host cell and enhance its reproduction. Though they may be altered by mutation, all of these genes except tev exist in all known variants of HIV; see Genetic variability of HIV.
HIV employs a sophisticated system of differential RNA splicing to obtain nine different gene products from a less than 10kb genome. HIV has a 9.2kb unspliced genomic transcript which encodes for gag and pol precursors; a singly spliced, 4.5 kb encoding for env, Vif, Vpr and Vpu and a multiply spliced, 2 kb mRNA encoding for Tat, Rev and Nef.
| Class | Gene name | Primary protein products | Processed protein products |
|---|---|---|---|
| Viral structural proteins | gag | Gag polyprotein | MA, CA, SP1, NC, SP2, P6 |
| pol | Pol polyprotein | RT, RNase H, IN, PR | |
| env | gp160 | gp120, gp41 | |
| Essential regulatory elements | tat | Tat |
|
| rev | Rev |
| |
| Accessory regulatory proteins | nef | Nef |
|
| vpr | Vpr |
| |
| vif | Vif |
| |
| vpu | Vpu |
|
Viral structural proteins
- gag (group-specific antigen) codes for the precursor gag polyprotein which is processed by viral protease during maturation to MA (matrix protein, p17); CA (capsid protein, p24); SP1 (spacer peptide 1, p2); NC (nucleocapsid protein, p7); SP2 (spacer peptide 2, p1) and P6 protein.
- pol codes for viral enzymes reverse transcriptase (RT) and RNase H, integrase (IN), and HIV protease (PR). HIV protease is required to cleave the precursor Gag polyprotein to produce structural proteins, RT is required to transcribe DNA from RNA template, and IN is necessary to integrate the double-stranded viral DNA into the host genome.
- env (for "envelope") codes for gp160, which is cleaved by a host protease, furin, within the endoplasmic reticulum of the host cell. The post-translational processing produces a surface glycoprotein, gp120 or SU, which attaches to the CD4 receptors present on lymphocytes, and gp41 or TM, which embeds in the viral envelope to enable the virus to attach to and fuse with target cells.
Essential regulatory elements
- tat (HIV trans-activator) plays an important role in regulating the reverse transcription of viral genome RNA, ensuring efficient synthesis of viral mRNAs and regulating the release of virions from infected cells. Tat is expressed as 72-amino acid one-exon Tat as well as the 86–101-amino-acid two-exon Tat, and plays an important role early in HIV infection. Tat (14–15 kDa) binds to the bulged genomic RNA stem-loop secondary structure near the 5' LTR region forming the trans-activation response element (TAR).
- rev (regulator of expression of virion proteins): The Rev protein binds to the viral genome via an arginine-rich RNA-binding motif that also acts as a NLS (nuclear localization signals), required for the transport of Rev to the nucleus from cytosol during viral replication. Rev recognizes a complex stem-loop structure of the mRNA env located in the intron separating coding exon of Tat and Rev, known as the HIV Rev response element (RRE). Rev is important for the synthesis of major viral proteins and is hence essential for viral replication.
Accessory regulatory proteins
- vpr (lentivirus protein R): Vpr is a virion-associated, nucleocytoplasmic shuttling regulatory protein. It is believed to play an important role in replication of the virus, specifically, nuclear import of the preintegration complex. Vpr also appears to cause its host cells to arrest their cell cycle in the G2 phase. This arrest activates the host DNA repair machinery which may enable integration of the viral DNA. HIV-2 and SIV encode an additional Vpr related protein called Vpx which functions in association with Vpr.
- vif - Vif is a highly conserved, 23 kDa phosphoprotein important for the infectivity of HIV-1 virions depending on the cell type. HIV-1 has been found to require Vif to synthesize infectious viruses in lymphocytes, macrophages, and certain human cell lines. It does not appear to require Vif for the same process in HeLa cells or COS cells, among others.
- nef- Nef, negative factor, is a N-terminal myristoylated membrane-associated phosphoprotein. It is involved in multiple functions during the replication cycle of the virus. It is believed to play an important role in cell apoptosis and increase virus infectivity.
- vpu (Virus protein U) - Vpu is specific to HIV-1. It is a class I oligomeric integral membrane phosphoprotein with numerous biological functions. Vpu is involved in CD4 degradation involving the ubiquitin proteasome pathway as well as in the successful release of virions from infected cells.
- tev: This gene is only present in a few HIV-1 isolates. It is a fusion of parts of the tat, env, and rev genes, and codes for a protein with some of the properties of tat, but little or none of the properties of rev.
RNA secondary structure
| HIV pol-1 stem loop | |
|---|---|
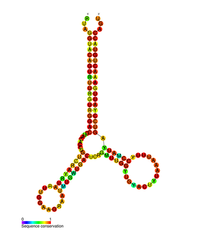 Predicted secondary structure of the HIV pol-1 stem loop | |
| Identifiers | |
| Symbol | pol |
| Rfam | RF01418 |
| Other data | |
| RNA type | Cis-reg |
| PDB structures | PDBe |
Several conserved secondary structure elements have been identified within the HIV RNA genome. The HIV viral RNA structures regulates the progression of reverse transcription. The 5'UTR structure consists of series of stem-loop structures connected by small linkers. These stem-loops (5' to 3') include the trans-activation region (TAR) element, the 5' polyadenylation signal [poly(A)], the PBS, the DIS, the major SD and the ψ hairpin structure located within the 5' end of the genome and the HIV Rev response element (RRE) within the env gene. Another RNA structure that has been identified is gag stem loop 3 (GSL3), thought to be involved in viral packaging. RNA secondary structures have been proposed to affect the HIV life cycle by altering the function of HIV protease and reverse transcriptase, although not all elements identified have been assigned a function.
An RNA secondary structure determined by SHAPE analysis has shown to contain three stem loops and is located between the HIV protease and reverse transcriptase genes. This cis regulatory RNA has been shown to be conserved throughout the HIV family and is thought to influence the viral life cycle.
V3 loop
The third variable loop or V3 loop is a part or region of the Human Immunodeficiency Virus. The V3 loop of the viron's envelope glycoprotein, gp120, allows it to infect human immune cells by binding to a cytokine receptor on the target human immune cell, such as a CCR5 cell or CXCR4 cell, depending on the strain of HIV. The envelope glycoprotein (Env) gp 120/41 is essential for HIV-1 entry into cells. Env serves as a molecular target of a medicine treating individuals with HIV-1 infection, and a source of immunogen to develop AIDS vaccine. However, the structure of the functional Env trimer has remained elusive.