Some chromosomal sex determination systems
A sex-determination system is a biological system that determines the development of sexual characteristics in an organism. Most organisms that create their offspring using sexual reproduction have two sexes. Occasionally, there are hermaphrodites in place of one or both sexes. There are also some species that are only one sex due to parthenogenesis, the act of a female reproducing without fertilization.
In many species, sex determination is genetic: males and females have different alleles or even different genes that specify their sexual morphology. In animals this is often accompanied by chromosomal differences, generally through combinations of XY, ZW, XO, ZO chromosomes, or haplodiploidy. The sexual differentiation is generally triggered by a main gene (a "sex locus"), with a multitude of other genes following in a domino effect.
In other cases, sex of a fetus is determined by environmental variables (such as temperature). The details of some sex-determination systems are not yet fully understood. Although, they do provide concrete analysis of complete biological sex-determinism. Hypothesis for future fetal biological system analysis include complete-reproduction-system initialized signals that can be measured during pregnancies to more accurately determine whether a determined sex of a fetus is male, or female. Such analysis of biological systems could also signal whether the fetus is hermaphrodite, which includes total or partial of both male and female reproduction organs.
Some species such as various flowers and fish do not have a fixed sex, and instead go through life cycles and change sex based on genetic cues during corresponding life stages of their type. This could be due to environmental factors such as seasons and temperature. Human fetus genitals can sometimes develop abnormalities during maternal pregnancies due to mutations in the fetuses sex-determinism system, resulting in the fetus becoming a hermaphrodite.
Chromosomal systems
XX/XY sex chromosomes
Drosophila sex-chromosomes
Human male XY chromosomes after G-banding
The XX/XY sex-determination system is the most familiar, as it is found in humans. The XX/XY system is found in most other mammals, as well as some insects. In this system, most females have two of the same kind of sex chromosome (XX), while most males have two distinct sex chromosomes (XY). The X and Y sex chromosomes are different in shape and size from each other, unlike the rest of the chromosomes (autosomes), and are sometimes called allosomes. In some species, such as humans, organisms remain sex indifferent for a time after they're created; in others, however, such as fruit flies, sexual differentiation occurs as soon as the egg is fertilized.[1]
Y-centered sex determination
Some species (including humans) have a gene SRY on the Y chromosome that determines maleness. Members of SRY-reliant species can have uncommon XY chromosomal combinations such as XXY and still live.[1] Human sex is determined by the presence or absence of a Y chromosome with a functional SRY gene. Once the SRY gene is activated, cells create testosterone and anti-müllerian hormone which typically ensures the development of a single, male reproductive system.[1] In typical XX embryos, cells secrete estrogen, which drives the body toward the female pathway.In Y-centered sex determination, the SRY gene is the main gene in determining male characteristics, but multiple genes are required to develop testes. In XY mice, lack of the gene DAX1 on the X chromosome results in sterility, but in humans it causes adrenal hypoplasia congenita.[2] However, when an extra DAX1 gene is placed on the X chromosome, the result is a female, despite the existence of SRY.[3] Even when there are normal sex chromosomes in XX females, duplication or expression of SOX9 causes testes to develop.[4][5] Gradual sex reversal in developed mice can also occur when the gene FOXL2 is removed from females.[6] Even though the gene DMRT1 is used by birds as their sex locus, species who have XY chromosomes also rely upon DMRT1, contained on chromosome 9, for sexual differentiation at some point in their formation.[1]
X-centered sex determination
Some species, such as fruit flies, use the presence of two X chromosomes to determine femaleness.[7] Species that use the number of Xs to determine sex are nonviable with an extra X chromosome.Other variants of XX/XY sex determination
Some fish have variants of the XY sex-determination system, as well as the regular system. For example, while having an XY format, Xiphophorus nezahualcoyotl and X. milleri also have a second Y chromosome, known as Y', that creates XY' females and YY' males.[8]At least one monotreme, the platypus, presents a particular sex determination scheme that in some ways resembles that of the ZW sex chromosomes of birds and lacks the SRY gene. The platypus has ten sex chromosomes; males have an XYXYXYXYXY pattern while females have ten X chromosomes. Although it is an XY system, the platypus' sex chromosomes share no homologues with eutherian sex chromosomes.[9] Instead, homologues with eutherian sex chromosomes lie on the platypus chromosome 6, which means that the eutherian sex chromosomes were autosomes at the time that the monotremes diverged from the therian mammals (marsupials and eutherian mammals). However, homologues to the avian DMRT1 gene on platypus sex chromosomes X3 and X5 suggest that it is possible the sex-determining gene for the platypus is the same one that is involved in bird sex-determination. More research must be conducted in order to determine the exact sex determining gene of the platypus.[10]
Heredity of sex chromosomes in XO sex determination
XX/X0 sex chromosomes
In this variant of the XY system, females have two copies of the sex chromosome (XX) but males have only one (X0). The 0 denotes the absence of a second sex chromosome. Generally in this method, the sex is determined by amount of genes expressed across the two chromosomes. This system is observed in a number of insects, including the grasshoppers and crickets of order Orthoptera and in cockroaches (order Blattodea). A small number of mammals also lack a Y chromosome. These include the Amami spiny rat (Tokudaia osimensis) and the Tokunoshima spiny rat (Tokudaia tokunoshimensis) and Sorex araneus, a shrew species. Transcaucasian mole voles (Ellobius lutescens) also have a form of XO determination, in which both sexes lack a second sex chromosome.[3] The mechanism of sex determination is not yet understood.[11]The nematode C. elegans is male with one sex chromosome (X0); with a pair of chromosomes (XX) it is a hermaphrodite.[12] Its main sex gene is XOL, which encodes XOL-1 and also controls the expression of the genes TRA-2 and HER-1. These genes reduce male gene activation and increase it, respectively.[13]
ZW sex chromosomes
The ZW sex-determination system is found in birds, some reptiles, and some insects and other organisms. The ZW sex-determination system is reversed compared to the XY system: females have two different kinds of chromosomes (ZW), and males have two of the same kind of chromosomes (ZZ). In the chicken, this was found to be dependent on the expression of DMRT1.[14] In birds, the genes FET1 and ASW are found on the W chromosome for females, similar to how the Y chromosome contains SRY.[1] However, not all species depend upon the W for their sex. For example, there are moths and butterflies that are ZW, but some have been found female with ZO, as well as female with ZZW.[12] Also, while mammals deactivate one of their extra X chromosomes when female, it appears that in the case of Lepidoptera, the males produce double the normal amount of enzymes, due to having two Z's.[12] Because the use of ZW sex determination is varied, it is still unknown how exactly most species determine their sex.[12] However, reportedly, the silkworm Bombyx mori uses a single female-specific piRNA as the primary determiner of sex.[15] Despite the similarities between the ZW and XY systems, these sex chromosomes evolved separately. In the case of the chicken, their Z chromosome is more similar to humans' autosome 9.[16] The chicken's Z chromosome also seems to be related to the X chromosome of the platypus.[17] When a ZW species, such as the Komodo dragon, reproduces parthenogenetically, usually only males are produced. This is due to the fact that the haploid eggs double their chromosomes, resulting in ZZ or WW. The ZZ become males, but the WW are not viable and are not brought to term.[18]UV sex chromosomes
In some Bryophyte and some algae species, the gametophyte stage of the life cycle, rather than being hermaphrodite, occurs as separate male or female individuals that produce male and female gametes respectively. When meiosis occurs in the sporophyte generation of the life cycle, the sex chromosomes known as U and V assort in spores that carry either the U chromosome and give rise to female gametophytes, or the V chromosome and give rise to male gametophytes.[19] [20]
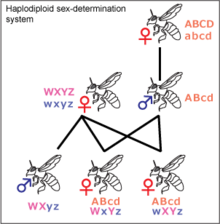
Haplodiploid sex chromosomes
Haplodiploidy
Haplodiploidy is found in insects belonging to Hymenoptera, such as ants and bees. Unfertilized eggs develop into haploid individuals, which are the males. Diploid individuals are generally female but may be sterile males. Males cannot have sons or fathers. If a queen bee mates with one drone, her daughters share ¾ of their genes with each other, not ½ as in the XY and ZW systems. This may be significant for the development of eusociality, as it increases the significance of kin selection, but it is debated.[21] Most females in the Hymenoptera order can decide the sex of their offspring by holding received sperm in their spermatheca and either releasing it into their oviduct or not. This allows them to create more workers, depending on the status of the colony.[22]Environmental systems
All alligators determine the sex of their offspring by the temperature of the nest.
Temperature-dependent
Many other sex-determination systems exist. In some species of reptiles, including alligators, some turtles, and the tuatara, sex is determined by the temperature at which the egg is incubated during a temperature-sensitive period. There are no examples of temperature-dependent sex determination (TSD) in birds. Megapodes had formerly been thought to exhibit this phenomenon, but were found to actually have different temperature-dependent embryo mortality rates for each sex.[23] For some species with TSD, sex determination is achieved by exposure to hotter temperatures resulting in the offspring being one sex and cooler temperatures resulting in the other. This type of TSD is called Pattern I. For others species using TSD, it is exposure to temperatures on both extremes that results in offspring of one sex, and exposure to moderate temperatures that results in offspring of the opposite sex, called Pattern II TSD. The specific temperatures required to produce each sex are known as the female-promoting temperature and the male-promoting temperature.[24] When the temperature stays near the threshold during the temperature sensitive period, the sex ratio is varied between the two sexes.[25] Some species' temperature standards are based on when a particular enzyme is created. These species that rely upon temperature for their sex determination do not have the SRY gene, but have other genes such as DAX1, DMRT1, and SOX9 that are expressed or not expressed depending on the temperature.[24] The sex of some species, such as the Nile tilapia, Australian skink lizard, and Australian dragon lizard, is initially determined by chromosomes, but can later be changed by the temperature of incubation.[8]It is unknown how exactly temperature-dependent sex determination evolved.[26] It could have evolved through certain sexes being more suited to certain areas that fit the temperature requirements. For example, a warmer area could be more suitable for nesting, so more females are produced to increase the amount that nest next season.[26] Environmental sex determination preceded the genetically determined systems of birds and mammals; it is thought that a temperature-dependent amniote was the common ancestor of amniotes with sex chromosomes.[citation needed]
Other systems
There are other environmental sex determination systems including location-dependent determination systems as seen in the marine worm Bonellia viridis – larvae become males if they make physical contact with a female, and females if they end up on the bare sea floor. This is triggered by the presence of a chemical produced by the females, bonellin.[27] Some species, such as some snails, practice sex change: adults start out male, then become female. In tropical clown fish, the dominant individual in a group becomes female while the other ones are male, and bluehead wrasses (Thalassoma bifasciatum) are the reverse. Some species, however, have no sex-determination system. Hermaphrodite species include the common earthworm and certain species of snails. A few species of fish, reptiles, and insects reproduce by parthenogenesis and are female altogether. There are some reptiles, such as the boa constrictor and Komodo dragon that can reproduce both sexually and asexually, depending on whether a mate is available.[28]Other unusual systems include those of the swordtail fish[clarification needed];[8] the Chironomus midges[clarification needed][citation needed]; the platypus, which has 10 sex chromosomes[9] but lacks the mammalian sex-determining gene SRY, meaning that the process of sex determination in the platypus remains unknown;[10] the juvenile hermaphroditism of zebrafish, with an unknown trigger;[8] and the platyfish, which has W, X, and Y chromosomes. This allows WY, WX, or XX females and YY or XY males.[8]
Evolution
Origin of sex chromosomes
The ends of the XY chromosomes, highlighted here in green, are all that is left of the original autosomes that can still cross-over with each other.
The accepted hypothesis of XY and ZW sex chromosome evolution is that they evolved at the same time, in two different branches.[29][30] However, there is some evidence to suggest that there could have been transitions between ZW and XY, such as in Xiphophorus maculatus, which have both ZW and XY systems in the same population, despite the fact that ZW and XY have different gene locations.[31][32] A recent theoretical model raises the possibility of both transitions between the XY/XX and ZZ/ZW system and environmental sex determination[33] The platypus' genes also back up the possible evolutionary link between XY and ZW, because they have the DMRT1 gene possessed by birds on their X chromosomes.[34] Regardless, XY and ZW follow a similar route. All sex chromosomes started out as an original autosome of an original amniote that relied upon temperature to determine the sex of offspring. After the mammals separated, the branch further split into Lepidosauria and Archosauromorpha. These two groups both evolved the ZW system separately, as evidenced by the existence of different sex chromosomal locations.[30] In mammals, one of the autosome pair, now Y, mutated its SOX3 gene into the SRY gene, causing that chromosome to designate sex.[30][34][35] After this mutation, the SRY-containing chromosome inverted and was no longer completely homologous with its partner. The regions of the X and Y chromosomes that are still homologous to one another are known as the pseudoautosomal region.[36] Once it inverted, the Y chromosome became unable to remedy deleterious mutations, and thus degenerated.[30] There is some concern that the Y chromosome will shrink further and stop functioning in ten million years: but the Y chromosome has been strictly conserved after its initial rapid gene loss.[37][38]
There are some species, such as the medaka fish, that evolved sex chromosomes separately; their Y chromosome never inverted and can still swap genes with the X. These species are still in an early phase of evolution with regard to their sex chromosomes. Because the Y does not have male-specific genes and can interact with the X, XY and YY females can be formed as well as XX males.[8]