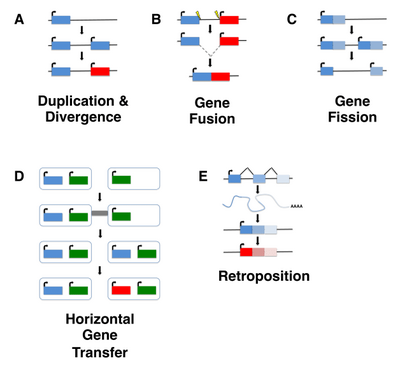
Homo erectus greatest extent (yellow),
Homo neanderthalensis greatest extent (ochre) and
Homo sapiens (red).

In paleoanthropology, the recent African origin of modern humans or the "Out of Africa" theory (OOA) is the most widely accepted model of the geographic origin and early migration of anatomically modern humans (Homo sapiens). It follows the early expansions of hominins out of Africa, accomplished by Homo erectus and then Homo neanderthalensis.
The model proposes a "single origin" of Homo sapiens in the taxonomic sense, precluding parallel evolution in other regions of traits considered anatomically modern, but not precluding multiple admixture between H. sapiens and archaic humans in Europe and Asia. H. sapiens most likely developed in the Horn of Africa between 300,000 and 200,000 years ago, although an alternative hypothesis argues that diverse morphological features of H. sapiens appeared locally in different parts of Africa and converged due to gene flow between different populations within the same period. The "recent African origin" model proposes that all modern non-African populations are substantially descended from populations of H. sapiens that left Africa after that time.
There were at least several "out-of-Africa" dispersals of modern humans, possibly beginning as early as 270,000 years ago, including 215,000 years ago to at least Greece, and certainly via northern Africa and the Arabian Peninsula about 130,000 to 115,000 years ago. There is evidence that modern humans had reached China around 80,000 years ago. Practically all of these early waves seem to have gone extinct or retreated back, and present-day humans outside Africa descend mainly from a single expansion out 70,000–50,000 years ago.
The most significant "recent" wave out of Africa took place about 70,000–50,000 years ago, via the so-called "Southern Route", spreading rapidly along the coast of Asia and reaching Australia by around 65,000–50,000 years ago, (though some researchers question the earlier Australian dates and place the arrival of humans there at 50,000 years ago at earliest, while others have suggested that these first settlers of Australia may represent an older wave before the more significant out of Africa migration and thus not necessarily be ancestral to the region's later inhabitants) while Europe was populated by an early offshoot which settled the Near East and Europe less than 55,000 years ago.
In the 2010s, studies in population genetics uncovered evidence of interbreeding that occurred between H. sapiens and archaic humans in Eurasia, Oceania and Africa, indicating that modern population groups, while mostly derived from early H. sapiens, are to a lesser extent also descended from regional variants of archaic humans.
Proposed waves
"Recent African origin", or Out of Africa II, refers to the migration of anatomically modern humans (Homo sapiens) out of Africa after their emergence at c. 300,000 to 200,000 years ago, in contrast to "Out of Africa I", which refers to the migration of archaic humans from Africa to Eurasia from before 1.8 and up to 0.5 million years ago. Omo-Kibish I (Omo I) from southern Ethiopia is the oldest anatomically modern Homo sapiens skeleton currently known (around 233,000 years old). There are even older Homo sapiens fossils from Jebel Irhoud in Morocco which exhibit a mixture of modern and archaic features at around 315,000 years old.
Since the beginning of the 21st century, the picture of "recent single-origin" migrations has become significantly more complex, due to the discovery of modern-archaic admixture and the increasing evidence that the "recent out-of-Africa" migration took place in waves over a long time. As of 2010, there were two main accepted dispersal routes for the out-of-Africa migration of early anatomically modern humans, the "Northern Route" (via Nile Valley and Sinai) and the "Southern Route" via the Bab-el-Mandeb strait.
- Posth et al. (2017) suggest that early Homo sapiens, or "another species in Africa closely related to us," might have first migrated out of Africa around 270,000 years ago based on the closer affinity within Neanderthals' mitochondrial genomes to Homo sapiens than Denisovans.
- Fossil evidence also points to an early Homo sapiens migration with the oldest known fossil coming from Apidima Cave in Greece and dated at 210,000 years ago. Finds at Misliya cave, which include a partial jawbone with eight teeth, have been dated to around 185,000 years ago. Layers dating from between 250,000 and 140,000 years ago in the same cave contained tools of the Levallois type which could put the date of the first migration even earlier if the tools can be associated with the modern human jawbone finds.
- An eastward dispersal from Northeast Africa to Arabia 150,000–130,000 years ago is based on the stone tools finds at Jebel Faya dated to 127,000 years ago (discovered in 2011), although fossil evidence in the area is significantly later at 85,000 years ago. Possibly related to this wave are the finds from Zhirendong cave, Southern China, dated to more than 100,000 years ago. Other evidence of modern human presence in China has been dated to 80,000 years ago.
- The most significant out of Africa dispersal took place around 50,000–70,000 years ago via the so-called Southern Route, either before or after the Toba event, which happened between 69,000 and 77,000 years ago. This dispersal followed the southern coastline of Asia and reached Australia around 65,000–50,000 years ago or according to some research, by 50,000 years ago at earliest. Western Asia was "re-occupied" by a different derivation from this wave around 50,000 years ago and Europe was populated from Western Asia beginning around 43,000 years ago.
- Wells (2003) describes an additional wave of migration after the southern coastal route, a northern migration into Europe about 45,000 years ago. This possibility is ruled out by Macaulay et al. (2005) and Posth et al. (2016), who argue for a single coastal dispersal, with an early offshoot into Europe.
Northern Route dispersal

Beginning 135,000 years ago, tropical Africa experienced megadroughts which drove humans from the land and towards the sea shores, and forced them to cross over to other continents.
Fossils of early Homo sapiens were found in Qafzeh and Es-Skhul Caves in Israel and have been dated to 80,000 to 120,000 years ago. These humans seem to have either become extinct or retreated back to Africa 70,000 to 80,000 years ago, possibly replaced by southbound Neanderthals escaping the colder regions of ice-age Europe. Hua Liu et al. analyzed autosomal microsatellite markers dating to about 56,000 years ago. They interpret the paleontological fossil as an isolated early offshoot that retracted back to Africa.
The discovery of stone tools in the United Arab Emirates in 2011 at the Faya-1 site in Mleiha, Sharjah, indicated the presence of modern humans at least 125,000 years ago, leading to a resurgence of the "long-neglected" North African route. This new understanding of the role of the Arabian dispersal began to change following results from archaeological and genetic studies stressing the importance of southern Arabia as a corridor for human expansions out of Africa.
In Oman, a site was discovered by Bien Joven in 2011 containing more than 100 surface scatters of stone tools belonging to the late Nubian Complex, known previously only from archaeological excavations in the Sudan. Two optically stimulated luminescence age estimates placed the Arabian Nubian Complex at approximately 106,000 years old. This provides evidence for a distinct Stone Age technocomplex in southern Arabia, around the earlier part of the Marine Isotope Stage 5.
According to Kuhlwilm and his co-authors, Neanderthals contributed genetically to modern humans then living outside of Africa around 100,000 years ago: humans which had already split off from other modern humans around 200,000 years ago, and this early wave of modern humans outside Africa also contributed genetically to the Altai Neanderthals. They found that "the ancestors of Neanderthals from the Altai Mountains and early modern humans met and interbred, possibly in the Near East, many thousands of years earlier than previously thought". According to co-author Ilan Gronau, "This actually complements archaeological evidence of the presence of early modern humans out of Africa around and before 100,000 years ago by providing the first genetic evidence of such populations." Similar genetic admixture events have been noted in other regions as well.
Southern Route dispersal
Coastal route
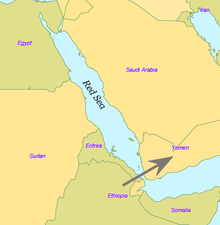
By some 50–70,000 years ago, a subset of the bearers of mitochondrial haplogroup L3 migrated from East Africa into the Near East. It has been estimated that from a population of 2,000 to 5,000 individuals in Africa, only a small group, possibly as few as 150 to 1,000 people, crossed the Red Sea. The group that crossed the Red Sea travelled along the coastal route around Arabia and the Persian Plateau to India, which appears to have been the first major settling point. Wells (2003) argued for the route along the southern coastline of Asia, across about 250 kilometres (155 mi), reaching Australia by around 50,000 years ago.

Today at the Bab-el-Mandeb straits, the Red Sea is about 20 kilometres (12 mi) wide, but 50,000 years ago sea levels were 70 m (230 ft) lower (owing to glaciation) and the water channel was much narrower. Though the straits were never completely closed, they were narrow enough to have enabled crossing using simple rafts, and there may have been islands in between. Shell middens 125,000 years old have been found in Eritrea, indicating that the diet of early humans included seafood obtained by beachcombing.
The dating of the Southern Dispersal is a matter of dispute. It may have happened either pre- or post-Toba, a catastrophic volcanic eruption that took place between 69,000 and 77,000 years ago at the site of present-day Lake Toba. Stone tools discovered below the layers of ash deposited in India may point to a pre-Toba dispersal but the source of the tools is disputed. An indication for post-Toba is haplo-group L3, that originated before the dispersal of humans out of Africa and can be dated to 60,000–70,000 years ago, "suggesting that humanity left Africa a few thousand years after Toba". Some research showing slower than expected genetic mutations in human DNA was published in 2012, indicating a revised dating for the migration to between 90,000 and 130,000 years ago. Some more recent research suggests a migration out-of-Africa of around 50,000-65,000 years ago of the ancestors of modern non-African populations, similar to most previous estimates.
Western Asia
Following the fossils dating 80,000 to 120,000 years ago from Qafzeh and Es-Skhul Caves in Israel there are no H. sapiens fossils in the Levant until the Manot 1 fossil from Manot Cave in Israel, dated to 54,700 years ago, though the dating was questioned by Groucutt et al. (2015). The lack of fossils and stone tool industries that can be safely associated with modern humans in the Levant has been taken to suggest that modern humans were outcompeted by Neanderthals until around 55,000 years ago, who would have placed a barrier on modern human dispersal out of Africa through the Northern Route. Climate reconstructions also support a Southern Route dispersal of modern humans as the Bab-el-Mandeb strait experienced a climate more conductive to human migration than the northern landbridge to the Levant during the major human dispersal out of Africa.
Oceania
It is thought that Australia was inhabited around 65,000–50,000 years ago. As of 2017, the earliest evidence of humans in Australia is at least 65,000 years old, while McChesney stated that
...genetic evidence suggests that a small band with the marker M168 migrated out of Africa along the coasts of the Arabian Peninsula and India, through Indonesia, and reached Australia very early, between 60,000 and 50,000 years ago. This very early migration into Australia is also supported by Rasmussen et al. (2011).
Fossils from Lake Mungo, Australia, have been dated to about 42,000 years ago. Other fossils from a site called Madjedbebe have been dated to at least 65,000 years ago, though some researchers doubt this early estimate and date the Madjedbebe fossils at about 50,000 years ago at the oldest.
Phylogenetic data suggests that an early Eastern Eurasian (Eastern non-African) meta-population trifurcated somewhere in eastern South Asia, and gave rise to the Australo-Papuans, the Ancient Ancestral South Indians (AASI), as well as East/Southeast Asians, although Papuans may have also received some gene flow from an earlier group (xOoA), around 2%, next to additional archaic admixture in the Sahul region.
According to one study, Papuans could have either formed from a mixture between an East Eurasian lineage and lineage basal to West and East Asians, or as a sister lineage of East Asians with or without a minor basal OoA or xOoA contribution.
A Holocene hunter-gatherer sample (Leang_Panninge) from South Sulawesi was found to be genetically in between East-Eurasians and Australo-Papuans. The sample could be modeled as ~50% Papuan-related and ~50% Basal-East Asian-related (Andamanese Onge or Tianyuan). The authors concluded that Basal-East Asian ancestry was far more widespread and the peopling of Insular Southeast Asia and Oceania was more complex than previously anticipated.


East and Southeast Asia
In China, the Liujiang man (Chinese: 柳江人) is among the earliest modern humans found in East Asia. The date most commonly attributed to the remains is 67,000 years ago. High rates of variability yielded by various dating techniques carried out by different researchers place the most widely accepted range of dates with 67,000 BP as a minimum, but do not rule out dates as old as 159,000 BP.[79] Liu, Martinón-Torres et al. (2015) claim that modern human teeth have been found in China dating to at least 80,000 years ago.
Tianyuan man from China has a probable date range between 38,000 and 42,000 years ago, while Liujiang man from the same region has a probable date range between 67,000 and 159,000 years ago. According to 2013 DNA tests, Tianyuan man is related "to many present-day Asians and Native Americans". Tianyuan is similar in morphology to Liujiang man, and some Jōmon period modern humans found in Japan, as well as modern East and Southeast Asians.
A 2021 study about the population history of Eastern Eurasia, concluded that distinctive Basal-East Asian (East-Eurasian) ancestry originated in Mainland Southeast Asia at ~50,000BC from a distinct southern Himalayan route, and expanded through multiple migration waves southwards and northwards respectively.
Genetic studies concluded that Native Americans descended from a single founding population that initially split from a Basal-East Asian source population in Mainland Southeast Asia around 36,000 years ago, at the same time at which the proper Jōmon people split from Basal-East Asians, either together with Ancestral Native Americans or during a separate expansion wave. They also show that the basal northern and southern Native American branches, to which all other Indigenous peoples belong, diverged around 16,000 years ago. An indigenous American sample from 16,000BC in Idaho, which is craniometrically similar to modern Native Americans as well as Paleosiberias, was found to have largely East-Eurasian ancestry and showed high affinity with contemporary East Asians, as well as Jōmon period samples of Japan, confirming that Ancestral Native Americans split from an East-Eurasian source population in Eastern Siberia.
Europe
According to Macaulay et al. (2005), an early offshoot from the southern dispersal with haplogroup N followed the Nile from East Africa, heading northwards and crossing into Asia through the Sinai. This group then branched, some moving into Europe and others heading east into Asia. This hypothesis is supported by the relatively late date of the arrival of modern humans in Europe as well as by archaeological and DNA evidence. Based on an analysis of 55 human mitochondrial genomes (mtDNAs) of hunter-gatherers, Posth et al. (2016) argue for a "rapid single dispersal of all non-Africans less than 55,000 years ago."
Genetic reconstruction
Mitochondrial haplogroups
Within Africa

The first lineage to branch off from Mitochondrial Eve was L0. This haplogroup is found in high proportions among the San of Southern Africa and the Sandawe of East Africa. It is also found among the Mbuti people. These groups branched off early in human history and have remained relatively genetically isolated since then. Haplogroups L1, L2, and L3 are descendants of L1–L6, and are largely confined to Africa. The macro haplogroups M and N, which are the lineages of the rest of the world outside Africa, descend from L3. L3 is about 70,000 years old, while haplogroups M and N are about 65–55,000 years old. The relationship between such gene trees and demographic history is still debated when applied to dispersals.
Of all the lineages present in Africa, the female descendants of only one lineage, mtDNA haplogroup L3, are found outside Africa. If there had been several migrations, one would expect descendants of more than one lineage to be found. L3's female descendants, the M and N haplogroup lineages, are found in very low frequencies in Africa (although haplogroup M1 populations are very ancient and diversified in North and North-east Africa) and appear to be more recent arrivals. A possible explanation is that these mutations occurred in East Africa shortly before the exodus and became the dominant haplogroups thereafter by means of the founder effect. Alternatively, the mutations may have arisen shortly afterwards.
Southern Route and haplogroups M and N

Results from mtDNA collected from aboriginal Malaysians called Orang Asli indicate that the haplogroups M and N share characteristics with original African groups from approximately 85,000 years ago, and share characteristics with sub-haplogroups found in coastal south-east Asian regions, such as Australasia, the Indian subcontinent and throughout continental Asia, which had dispersed and separated from their African progenitor approximately 65,000 years ago. This southern coastal dispersal would have occurred before the dispersal through the Levant approximately 45,000 years ago. This hypothesis attempts to explain why haplogroup N is predominant in Europe and why haplogroup M is absent in Europe. Evidence of the coastal migration is thought to have been destroyed by the rise in sea levels during the Holocene epoch. Alternatively, a small European founder population that had expressed haplogroup M and N at first, could have lost haplogroup M through random genetic drift resulting from a bottleneck (i.e. a founder effect).
The group that crossed the Red Sea travelled along the coastal route around Arabia and Persia until reaching India. Haplogroup M is found in high frequencies along the southern coastal regions of Pakistan and India and it has the greatest diversity in India, indicating that it is here where the mutation may have occurred. Sixty percent of the Indian population belong to Haplogroup M. The indigenous people of the Andaman Islands also belong to the M lineage. The Andamanese are thought to be offshoots of some of the earliest inhabitants in Asia because of their long isolation from the mainland. They are evidence of the coastal route of early settlers that extends from India to Thailand and Indonesia all the way to eastern New Guinea. Since M is found in high frequencies in highlanders from New Guinea and the Andamanese and New Guineans have dark skin and Afro-textured hair, some scientists think they are all part of the same wave of migrants who departed across the Red Sea ~60,000 years ago in the Great Coastal Migration. The proportion of haplogroup M increases eastwards from Arabia to India; in eastern India, M outnumbers N by a ratio of 3:1. Crossing into Southeast Asia, haplogroup N (mostly in the form of derivatives of its R subclade) reappears as the predominant lineage. M is predominant in East Asia, but amongst Indigenous Australians, N is the more common lineage. This haphazard distribution of Haplogroup N from Europe to Australia can be explained by founder effects and population bottlenecks.

Autosomal DNA
A 2002 study of African, European, and Asian populations, found greater genetic diversity among Africans than among Eurasians, and that genetic diversity among Eurasians is largely a subset of that among Africans, supporting the out of Africa model. A large study by Coop et al. (2009) found evidence for natural selection in autosomal DNA outside of Africa. The study distinguishes non-African sweeps (notably KITLG variants associated with skin color), West-Eurasian sweeps (SLC24A5) and East-Asian sweeps (MC1R, relevant to skin color). Based on this evidence, the study concluded that human populations encountered novel selective pressures as they expanded out of Africa. MC1R and its relation to skin color had already been discussed by Harding et al. (2000), p. 1355. According to this study, Papua New Guineans continued to be exposed to selection for dark skin color so that, although these groups are distinct from Africans in other places, the allele for dark skin color shared by contemporary Africans, Andamanese and New Guineans is an archaism. Endicott et al. (2003) suggest convergent evolution. A 2014 study by Gurdasani et al. indicates that the higher genetic diversity in Africa was further increased in some regions by relatively recent Eurasian migrations affecting parts of Africa.
Pathogen DNA
Another promising route towards reconstructing human genetic genealogy is via the JC virus (JCV), a type of human polyomavirus which is carried by 70–90 percent of humans and which is usually transmitted vertically, from parents to offspring, suggesting codivergence with human populations. For this reason, JCV has been used as a genetic marker for human evolution and migration. This method does not appear to be reliable for the migration out of Africa; in contrast to human genetics, JCV strains associated with African populations are not basal. From this Shackelton et al. (2006) conclude that either a basal African strain of JCV has become extinct or that the original infection with JCV post-dates the migration from Africa.
Admixture of archaic and modern humans
Evidence for archaic human species (descended from Homo heidelbergensis) having interbred with modern humans outside of Africa, was discovered in the 2010s. This concerns primarily Neanderthal admixture in all modern populations except for Sub-Saharan Africans but evidence has also been presented for Denisova hominin admixture in Australasia (i.e. in Melanesians, Aboriginal Australians and some Negritos). The rate of Neanderthal admixture to European and Asian populations as of 2017 has been estimated at between about 2–3%.
Archaic admixture in some Sub-Saharan African populations hunter-gatherer groups (Biaka Pygmies and San), derived from archaic hominins that broke away from the modern human lineage around 700,000 years ago, was discovered in 2011. The rate of admixture was estimated at 2%. Admixture from archaic hominins of still earlier divergence times, estimated at 1.2 to 1.3 million years ago, was found in Pygmies, Hadza and five Sandawe in 2012.
From an analysis of Mucin 7, a highly divergent haplotype that has an estimated coalescence time with other variants around 4.5 million years BP and is specific to African populations, it is inferred to have been derived from interbreeding between African modern and archaic humans.
A study published in 2020 found that the Yoruba and Mende populations of West Africa derive between 2% and 19% of their genome from an as-yet unidentified archaic hominin population that likely diverged before the split of modern humans and the ancestors of Neanderthals and Denisovans.
Stone tools
In addition to genetic analysis, Petraglia et al. also examines the small stone tools (microlithic materials) from the Indian subcontinent and explains the expansion of population based on the reconstruction of paleoenvironment. He proposed that the stone tools could be dated to 35 ka in South Asia, and the new technology might be influenced by environmental change and population pressure.
History of the theory
Classical paleoanthropology

The cladistic relationship of humans with the African apes was suggested by Charles Darwin after studying the behaviour of African apes, one of which was displayed at the London Zoo. The anatomist Thomas Huxley had also supported the hypothesis and suggested that African apes have a close evolutionary relationship with humans. These views were opposed by the German biologist Ernst Haeckel, who was a proponent of the Out of Asia theory. Haeckel argued that humans were more closely related to the primates of South-east Asia and rejected Darwin's African hypothesis.
In the Descent of Man, Darwin speculated that humans had descended from apes, which still had small brains but walked upright, freeing their hands for uses which favoured intelligence; he thought such apes were African:
In each great region of the world the living mammals are closely related to the extinct species of the same region. It is, therefore, probable that Africa was formerly inhabited by extinct apes closely allied to the gorilla and chimpanzee; and as these two species are now man's nearest allies, it is somewhat more probable that our early progenitors lived on the African continent than elsewhere. But it is useless to speculate on this subject, for an ape nearly as large as a man, namely the Dryopithecus of Lartet, which was closely allied to the anthropomorphous Hylobates, existed in Europe during the Upper Miocene period; and since so remote a period the earth has certainly undergone many great revolutions, and there has been ample time for migration on the largest scale.
— Charles Darwin, Descent of Man
In 1871, there were hardly any human fossils of ancient hominins available. Almost fifty years later, Darwin's speculation was supported when anthropologists began finding fossils of ancient small-brained hominins in several areas of Africa (list of hominina fossils). The hypothesis of recent (as opposed to archaic) African origin developed in the 20th century. The "Recent African origin" of modern humans means "single origin" (monogenism) and has been used in various contexts as an antonym to polygenism. The debate in anthropology had swung in favour of monogenism by the mid-20th century. Isolated proponents of polygenism held forth in the mid-20th century, such as Carleton Coon, who thought as late as 1962 that H. sapiens arose five times from H. erectus in five places.

a: Exit of the L3 precursor to Eurasia. b: Return to Africa and expansion to Asia of basal L3 lineages with subsequent differentiation in both continents.
Multiregional origin hypothesis
The historical alternative to the recent origin model is the multiregional origin of modern humans, initially proposed by Milford Wolpoff in the 1980s. This view proposes that the derivation of anatomically modern human populations from H. erectus at the beginning of the Pleistocene 1.8 million years BP, has taken place within a continuous world population. The hypothesis necessarily rejects the assumption of an infertility barrier between ancient Eurasian and African populations of Homo. The hypothesis was controversially debated during the late 1980s and the 1990s. The now-current terminology of "recent-origin" and "Out of Africa" became current in the context of this debate in the 1990s. Originally seen as an antithetical alternative to the recent origin model, the multiregional hypothesis in its original "strong" form is obsolete, while its various modified weaker variants have become variants of a view of "recent origin" combined with archaic admixture. Stringer (2014) distinguishes the original or "classic" Multiregional model as having existed from 1984 (its formulation) until 2003, to a "weak" post-2003 variant that has "shifted close to that of the Assimilation Model".
Mitochondrial analyses
In the 1980s, Allan Wilson together with Rebecca L. Cann and Mark Stoneking worked on genetic dating of the matrilineal most recent common ancestor of modern human populations (dubbed "Mitochondrial Eve"). To identify informative genetic markers for tracking human evolutionary history, Wilson concentrated on mitochondrial DNA (mtDNA), passed from mother to child. This DNA material mutates quickly, making it easy to plot changes over relatively short times. With his discovery that human mtDNA is genetically much less diverse than chimpanzee mtDNA, Wilson concluded that modern human populations had diverged recently from a single population while older human species such as Neanderthals and Homo erectus had become extinct. With the advent of archaeogenetics in the 1990s, the dating of mitochondrial and Y-chromosomal haplogroups became possible with some confidence. By 1999, estimates ranged around 150,000 years for the mt-MRCA and 60,000 to 70,000 years for the migration out of Africa.
From 2000 to 2003, there was controversy about the mitochondrial DNA of "Mungo Man 3" (LM3) and its possible bearing on the multiregional hypothesis. LM3 was found to have more than the expected number of sequence differences when compared to modern human DNA (CRS). Comparison of the mitochondrial DNA with that of ancient and modern aborigines, led to the conclusion that Mungo Man fell outside the range of genetic variation seen in Aboriginal Australians and was used to support the multiregional origin hypothesis. A reanalysis of LM3 and other ancient specimens from the area published in 2016, showed it to be akin to modern Aboriginal Australian sequences, inconsistent with the results of the earlier study.
Y-chromosome analyses
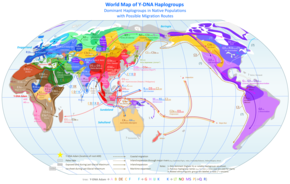
As current estimates on the male most recent common ancestor ("Y-chromosomal Adam" or Y-MRCA) converge with estimates for the age of anatomically modern humans, and well predate the Out of Africa migration, geographical origin hypotheses continue to be limited to the African continent.
The most basal lineages have been detected in West, Northwest and Central Africa, suggesting plausibility for the Y-MRCA living in the general region of "Central-Northwest Africa".
Another study finds a plausible placement in "the north-western quadrant of the African continent" for the emergence of the A1b haplogroup. The 2013 report of haplogroup A00 found among the Mbo people of western present-day Cameroon is also compatible with this picture.
The revision of Y-chromosomal phylogeny since 2011 has affected estimates for the likely geographical origin of Y-MRCA as well as estimates on time depth. By the same reasoning, future discovery of presently-unknown archaic haplogroups in living people would again lead to such revisions. In particular, the possible presence of between 1% and 4% Neanderthal-derived DNA in Eurasian genomes implies that the (unlikely) event of a discovery of a single living Eurasian male exhibiting a Neanderthal patrilineal line would immediately push back T-MRCA ("time to MRCA") to at least twice its current estimate. However, the discovery of a Neanderthal Y-chromosome by Mendez et al. was tempered by a 2016 study that suggests the extinction of Neanderthal patrilineages, as the lineage inferred from the Neanderthal sequence is outside of the range of contemporary human genetic variation. Questions of geographical origin would become part of the debate on Neanderthal evolution from Homo erectus.