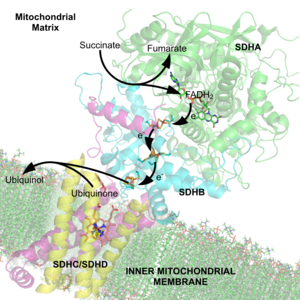
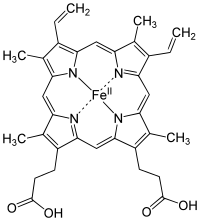
The succinate dehydrogenase complex showing several cofactors, including flavin, iron-sulfur centers, and heme.
A cofactor is a non-protein chemical compound or metallic ion that is required for an enzyme's activity. Cofactors can be considered "helper molecules" that assist in biochemical transformations. The rates at which these happen are characterized by enzyme kinetics.
Cofactors can be subclassified as either inorganic ions or complex organic molecules called coenzymes, the latter of which is mostly derived from vitamins and other organic essential nutrients in small amounts. A coenzyme that is tightly or even covalently bound is termed a prosthetic group. Cosubstrates
are transiently bound to the protein and will be released at some
point, then get back in. The prosthetic groups, on the other hand, are
bound permanently to the protein. Both of them have the same function,
which is to facilitate the reaction of enzymes and protein.
Additionally, some sources also limit the use of the term "cofactor" to
inorganic substances. An inactive enzyme without the cofactor is called an apoenzyme, while the complete enzyme with cofactor is called a holoenzyme.
Some enzymes or enzyme complexes require several cofactors. For example, the multienzyme complex pyruvate dehydrogenase at the junction of glycolysis and the citric acid cycle requires five organic cofactors and one metal ion: loosely bound thiamine pyrophosphate (TPP), covalently bound lipoamide and flavin adenine dinucleotide (FAD), and the cosubstrates nicotinamide adenine dinucleotide (NAD+) and coenzyme A (CoA), and a metal ion (Mg2+).
Organic cofactors are often vitamins or made from vitamins. Many contain the nucleotide adenosine monophosphate (AMP) as part of their structures, such as ATP, coenzyme A, FAD, and NAD+. This common structure may reflect a common evolutionary origin as part of ribozymes in an ancient RNA world.
It has been suggested that the AMP part of the molecule can be
considered to be a kind of "handle" by which the enzyme can "grasp" the
coenzyme to switch it between different catalytic centers.
Classification
Cofactors can be divided into two major groups: organic Cofactors, such as flavin or heme, and inorganic cofactors, such as the metal ions Mg2+, Cu+, Mn2+, or iron-sulfur clusters.
Organic cofactors are sometimes further divided into coenzymes and prosthetic groups.
The term coenzyme refers specifically to enzymes and, as such, to the
functional properties of a protein. On the other hand, "prosthetic
group" emphasizes the nature of the binding of a cofactor to a protein
(tight or covalent) and, thus, refers to a structural property.
Different sources give slightly different definitions of coenzymes,
cofactors, and prosthetic groups. Some consider tightly bound organic
molecules as prosthetic groups and not as coenzymes, while others define
all non-protein organic molecules needed for enzyme activity as
coenzymes, and classify those that are tightly bound as coenzyme
prosthetic groups. These terms are often used loosely.
A 1980 letter in Trends in Biochemistry Sciences noted the
confusion in the literature and the essentially arbitrary distinction
made between prosthetic groups and coenzymes group and proposed the
following scheme. Here, cofactors were defined as an additional
substance apart from protein and substrate that is required for enzyme activity and a prosthetic group as a substance that undergoes its whole catalytic cycle
attached to a single enzyme molecule. However, the author could not
arrive at a single all-encompassing definition of a "coenzyme" and
proposed that this term be dropped from use in the literature.
Inorganic
Metal ions
Metal ions are common cofactors. The study of these cofactors falls under the area of bioinorganic chemistry. In nutrition, the list of essential trace elements reflects their role as cofactors. In humans this list commonly includes iron, magnesium, manganese, cobalt, copper, zinc, and molybdenum. Although chromium deficiency causes impaired glucose tolerance, no human enzyme that uses this metal as a cofactor has been identified. Iodine is also an essential trace element, but this element is used as part of the structure of thyroid hormones rather than as an enzyme cofactor. Calcium
is another special case, in that it is required as a component of the
human diet, and it is needed for the full activity of many enzymes, such
as nitric oxide synthase, protein phosphatases, and adenylate kinase, but calcium activates these enzymes in allosteric regulation, often binding to these enzymes in a complex with calmodulin. Calcium is, therefore, a cell signaling molecule, and not usually considered a cofactor of the enzymes it regulates.
Other organisms require additional metals as enzyme cofactors, such as vanadium in the nitrogenase of the nitrogen-fixing bacteria of the genus Azotobacter, tungsten in the aldehyde ferredoxin oxidoreductase of the thermophilic archaean Pyrococcus furiosus, and even cadmium in the carbonic anhydrase from the marine diatom Thalassiosira weissflogii.
In many cases, the cofactor includes both an inorganic and organic component. One diverse set of examples is the heme proteins, which consist of a porphyrin ring coordinated to iron.
A simple [Fe2S2] cluster containing two iron atoms and two sulfur atoms, coordinated by four protein cysteine residues.
Iron-sulfur clusters
Iron-sulfur clusters are complexes of iron and sulfur atoms held
within proteins by cysteinyl residues. They play both structural and
functional roles, including electron transfer, redox sensing, and as
structural modules.
Organic
Organic
cofactors are small organic molecules (typically a molecular mass less
than 1000 Da) that can be either loosely or tightly bound to the enzyme
and directly participate in the reaction. In the latter case, when it is difficult to remove without denaturing the enzyme, it can be called a prosthetic group. It is important to emphasize that there is no sharp division between loosely and tightly bound cofactors. Indeed, many such as NAD+ can be tightly bound in some enzymes, while it is loosely bound in others. Another example is thiamine pyrophosphate (TPP), which is tightly bound in transketolase or pyruvate decarboxylase, while it is less tightly bound in pyruvate dehydrogenase. Other coenzymes, flavin adenine dinucleotide (FAD), biotin, and lipoamide, for instance, are tightly bound.
Tightly bound cofactors are, in general, regenerated during the same
reaction cycle, while loosely bound cofactors can be regenerated in a
subsequent reaction catalyzed by a different enzyme. In the latter case,
the cofactor can also be considered a substrate or cosubstrate.
Vitamins can serve as precursors to many organic cofactors (e.g., vitamins B1, B2, B6, B12, niacin, folic acid) or as coenzymes themselves (e.g., vitamin C). However, vitamins do have other functions in the body. Many organic cofactors also contain a nucleotide, such as the electron carriers NAD and FAD, and coenzyme A, which carries acyl
groups. Most of these cofactors are found in a huge variety of species,
and some are universal to all forms of life. An exception to this wide
distribution is a group of unique cofactors that evolved in methanogens, which are restricted to this group of archaea.
Vitamins and derivatives
Non-vitamins
| Cofactor | Chemical group(s) transferred | Distribution |
| Adenosine triphosphate | Phosphate group | Bacteria, archaea,and eukaryotes |
| S-Adenosyl methionine | Methyl group | Bacteria, archaea. and eukaryotes |
| Coenzyme B | Electrons | Methanogens |
| Coenzyme M | Methyl group | Methanogens |
| Coenzyme Q | Electrons | Bacteria, archaea,and eukaryotes |
| Cytidine triphosphate | Diacylglycerols and lipid head groups | Bacteria, archaea,and eukaryotes |
| Glutathione | Electrons | Some bacteriaand most eukaryotes |
| Heme | Electrons | Bacteria, archaeaand eukaryotes |
| Lipoamide | Electrons, acyl groups | Bacteria, archaeaand eukaryotes |
| Methanofuran | Formyl group | Methanogens |
| Molybdopterin | Oxygen atoms | Bacteria, archaea and eukaryotes |
| Nucleotide sugars | Monosaccharides | Bacteria, archaeaand eukaryotes |
| 3'-Phosphoadenosine-5'-phosphosulfate | Sulfate group | Bacteria, archaeaand eukaryotes |
| Pyrroloquinoline quinone | Electrons | Bacteria |
| Tetrahydrobiopterin | Oxygen atom and electrons | Bacteria, archaeaand eukaryotes |
| Tetrahydromethanopterin | Methyl group | Methanogens |
Cofactors as metabolic intermediates
The redox reactions of nicotinamide adenine dinucleotide.
Metabolism involves a vast array of chemical reactions, but most fall
under a few basic types of reactions that involve the transfer of functional groups.
This common chemistry allows cells to use a small set of metabolic
intermediates to carry chemical groups between different reactions. These group-transfer intermediates are the loosely bound organic cofactors, often called coenzymes.
Each class of group-transfer reaction is carried out by a
particular cofactor, which is the substrate for a set of enzymes that
produce it, and a set of enzymes that consume it. An example of this are
the dehydrogenases that use nicotinamide adenine dinucleotide (NAD+) as a cofactor. Here, hundreds of separate types of enzymes remove electrons from their substrates and reduce NAD+ to NADH. This reduced cofactor is then a substrate for any of the reductases in the cell that require electrons to reduce their substrates.
Therefore, these cofactors are continuously recycled as part of metabolism. As an example, the total quantity of ATP in the human body is about 0.1 mole.
This ATP is constantly being broken down into ADP, and then converted
back into ATP. Thus, at any given time, the total amount of ATP + ADP
remains fairly constant. The energy used by human cells requires the hydrolysis
of 100 to 150 moles of ATP daily, which is around 50 to 75 kg. In
typical situations, humans use up their body weight of ATP over the
course of the day. This means that each ATP molecule is recycled 1000 to 1500 times daily.
Evolution
Organic cofactors, such as ATP and NADH, are present in all known forms of life and form a core part of metabolism. Such universal conservation indicates that these molecules evolved very early in the development of living things. At least some of the current set of cofactors may, therefore, have been present in the last universal ancestor, which lived about 4 billion years ago.
Organic cofactors may have been present even earlier in the history of life on Earth. The nucleotide adenosine
is present in cofactors that catalyse many basic metabolic reactions
such as methyl, acyl, and phosphoryl group transfer, as well as redox reactions. This ubiquitous chemical scaffold has, therefore, been proposed to be a remnant of the RNA world, with early ribozymes evolving to bind a restricted set of nucleotides and related compounds.
Adenosine-based cofactors are thought to have acted as interchangeable
adaptors that allowed enzymes and ribozymes to bind new cofactors
through small modifications in existing adenosine-binding domains, which had originally evolved to bind a different cofactor. This process of adapting a pre-evolved structure for a novel use is known as exaptation.
A computational method, IPRO, recently predicted mutations that
experimentally switched the cofactor specificity of Candida boidinii
xylose reductase from NADPH to NADH.
History
The first organic cofactor to be discovered was NAD+, which was identified by Arthur Harden and William Youndin 1906. They noticed that adding boiled and filtered yeast extract greatly accelerated alcoholic fermentation in unboiled yeast extracts. They called the unidentified factor responsible for this effect a coferment. Through a long and difficult purification from yeast extracts, this heat-stable factor was identified as a nucleotide sugar phosphate by Hans von Euler-Chelpin. Other cofactors were identified throughout the early 20th century, with ATP being isolated in 1929 by Karl Lohmann, and coenzyme A being discovered in 1945 by Fritz Albert Lipmann.
The functions of these molecules were at first mysterious, but, in 1936, Otto Heinrich Warburg identified the function of NAD+ in hydride transfer. This discovery was followed in the early 1940s by the work of Herman Kalckar, who established the link between the oxidation of sugars and the generation of ATP. This confirmed the central role of ATP in energy transfer that had been proposed by Fritz Albert Lipmann in 1941. Later, in 1949, Morris Friedkin and Albert L. Lehninger proved that NAD+ linked metabolic pathways such as the citric acid cycle and the synthesis of ATP.
Protein-derived cofactors
In
a number of enzymes, the moiety that acts as a cofactor is formed by
post-translational modification of a part of the protein sequence. This
often replaces the need for an external binding factor, such as a metal
ion, for protein function. Potential modifications could be oxidation
of aromatic residues, binding between residues, cleavage or
ring-forming. These alterations are distinct from other post-translation protein modifications, such as phosphorylation,
methylation, or glycosylation in that the amino acids typically acquire
new functions. This increases the functionality of the protein;
unmodified amino acids are typically limited to acid-base reactions, and
the alteration of resides can give the protein electrophilic sites or
the ability to stabilize free radicals. Examples of cofactor production include tryptophan tryptophylquinone (TTQ), derived from two tryptophan side chains, and 4-methylidene-imidazole-5-one (MIO), derived from an Ala-Ser-Gly motif. Characterization of protein-derived cofactors is conducted using X-ray crystallography and mass spectroscopy; structural data is necessary because sequencing does not readily identify the altered sites.
Non-enzymatic cofactors
The
term is used in other areas of biology to refer more broadly to
non-protein (or even protein) molecules that either activate, inhibit,
or are required for the protein to function. For example, ligands such as hormones that bind to and activate receptor proteins
are termed cofactors or coactivators, whereas molecules that inhibit
receptor proteins are termed corepressors. One such example is the G
protein-coupled receptor family of receptors, which are frequently found
in sensory neurons. Ligand binding to the receptors activates the G
protein, which then activates an enzyme to activate the effector.
In order to avoid confusion, it has been suggested that such proteins
that have ligand-binding mediated activation or repression be referred
to as coregulators.