From Wikipedia, the free encyclopedia
Antibiotic
resistance tests: Bacteria are streaked on dishes with white disks,
each impregnated with a different antibiotic. Clear rings, such as those
on the left, show that bacteria have not grown—indicating that these
bacteria are not resistant. The bacteria on the right are fully
susceptible to only three of the seven antibiotics tested.
[1]
Antimicrobial resistance (
AMR or
AR) is the ability of a microbe to resist the effects of medication that once could successfully treat the microbe. The term
antibiotic resistance (
AR or
ABR) is a subset of AMR, as it applies only to
bacteria becoming resistant to
antibiotics.
Resistant microbes are more difficult to treat, requiring alternative
medications or higher doses of antimicrobials. These approaches may be
more expensive,
more toxic or both. Microbes resistant to multiple antimicrobials are called
multidrug resistant (MDR). Those considered extensively drug resistant (XDR) or totally drug resistant (TDR) are sometimes called "superbugs".
Resistance arises through one of three mechanisms: natural resistance in certain types of bacteria, genetic
mutation, or by one species acquiring resistance from another.
[6] All classes of microbes can develop resistance. Fungi develop
antifungal resistance.
Viruses develop
antiviral resistance.
Protozoa develop
antiprotozoal resistance, and
bacteria develop
antibiotic
resistance. Resistance can appear spontaneously because of random
mutations. However, extended use of antimicrobials appears to encourage
mutations which can render antimicrobials ineffective.
[7]
Preventive measures include only using antibiotics when needed, thereby stopping
misuse of antibiotics or antimicrobials.
[8][9]
Narrow-spectrum antibiotics are preferred over broad-spectrum
antibiotics when possible, as effectively and accurately targeting
specific organisms is less likely to cause resistance.
[10]
For people who take these medications at home, education about proper
use is essential. Health care providers can minimize spread of resistant
infections by use of proper
sanitation and
hygiene, including
handwashing and disinfecting between patients, and should encourage the same of the patient, visitors, and family members.
[11]
Rising drug resistance is caused mainly by use of antimicrobials
in humans and other animals, and spread of resistant strains between the
two.
[8]
Growing resistance has also been linked to dumping of inadequately
treated effluents from the pharmaceutical industry, especially in
countries where bulk drugs are manufactured.
[12] Antibiotics increase
selective pressure
in bacterial populations, causing vulnerable bacteria to die; this
increases the percentage of resistant bacteria which continue growing.
Even at very low levels of antibiotic, resistant bacteria can have a
growth advantage and grow faster than vulnerable bacteria.
[13]
With resistance to antibiotics becoming more common there is greater
need for alternative treatments. Calls for new antibiotic therapies have
been issued, but new drug development is becoming rarer.
[14]
Antimicrobial resistance is increasing globally because of greater access to antibiotic drugs in
developing countries.
[15] Estimates are that 700,000 to several million deaths result per year.
[16][17]
Each year in the United States, at least 2 million people become
infected with bacteria that are resistant to antibiotics and at least
23,000 people die as a result.
[18] There are public calls for global collective action to address the threat that include proposals for
international treaties on antimicrobial resistance.
[19]
Worldwide antibiotic resistance is not completely identified, but
poorer countries with weaker healthcare systems are more affected.
[9]
Definition
Diagram
showing the difference between non-resistant bacteria and drug
resistant bacteria. Non-resistant bacteria multiply, and upon drug
treatment, the bacteria die. Drug resistant bacteria multiply as well,
but upon drug treatment, the bacteria continue to spread.
[20]
The WHO defines antimicrobial resistance as a microorganism's
resistance to an antimicrobial drug that was once able to treat an
infection by that microorganism.
[3]
A person cannot become resistant to antibiotics. Resistance is a
property of the microbe, not a person or other organism infected by a
microbe.
[21]
Overview
A
World Health Organization
(WHO) report released April 2014 stated, "this serious threat is no
longer a prediction for the future, it is happening right now in every
region of the world and has the potential to affect anyone, of any age,
in any country. Antibiotic resistance—when bacteria change so
antibiotics no longer work in people who need them to treat
infections—is now a major threat to public health."
[22]
Causes
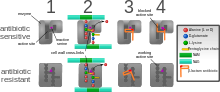
How antibiotic resistance evolves and spreads
Bacteria with resistance to antibiotics predate medical use of antibiotics by humans.
[23] However, widespread antibiotic use has made more bacteria resistant through the process of
evolutionary pressure.
[7]
Reasons for the widespread use of antibiotics in human medicine include:
- increasing global availability over time since the 1950s
- uncontrolled sale in many low or middle income countries, where they
can be obtained over the counter without a prescription, potentially
resulting in antibiotics being used when not indicated.[26]:1060 This may result in emergence of resistance in any remaining bacteria.
Other causes include:
- Antibiotic use in livestock
feed at low doses for growth promotion is an accepted practice in many
industrialized countries and is known to lead to increased levels of
resistance.[27][28]
- Releasing large quantities of antibiotics into the environment during pharmaceutical manufacturing through inadequate wastewater treatment increases the risk that antibiotic-resistant strains will develop and spread.[29][30]
- It is uncertain whether antibacterials in soaps and other products contribute to antibiotic resistance, but antibacterial soaps are discouraged for other reasons.[31][32]
Human medicine
Deaths attributable to antimicrobial resistance every year compared to other major causes of death.
[17]
Increasing bacterial resistance is linked with the volume of
antibiotic prescribed, as well as missing doses when taking antibiotics.
[33]
Inappropriate prescribing of antibiotics has been attributed to a
number of causes, such as patients insisting on antibiotics and
physicians prescribing them as they do not have time to explain why they
are not necessary. Another cause can be physicians not knowing when to
prescribe antibiotics or being overly cautious for medical or legal
reasons.
[34] For example, 70 to 80 percent of
diarrhea
is caused by viral pathogens, for which antibiotics are not effective.
But nevertheless, around 40 percent of these cases are attempted to be
treated with antibiotics.
[35] In some areas even over 80 percent of such cases are attempted to be treated with antibiotics.
[35]
Lower antibiotic concentration contributes to the increase of AMR
by introducing more mutations that support bacterial growth in higher
antibiotic concentration. For example, sub-inhibitory concentration have
induced genetic mutation in bacteria such as
Pseudomonas aeruginosa and
Bacteroides fragilis.
[36]
Up to half of antibiotics used in humans are unnecessary and inappropriate.
[8] For example, a third of people believe that antibiotics are effective for the
common cold,
[37] and the common cold is the most common reason antibiotics are prescribed even though antibiotics are useless against viruses.
[38]
A single regimen of antibiotics even in compliant individuals leads to a
greater risk of resistant organisms to that antibiotic in the person
for a month to possibly a year.
[39][40]
Antibiotic resistance increases with duration of treatment.
Therefore, as long as an effective minimum is kept, shorter courses of
antibiotics are likely to decrease rates of resistance, reduce cost, and
have better outcomes with fewer complications.
[10] Short course regimens exist for
community-acquired pneumonia[41] spontaneous bacterial peritonitis,
[42] suspected lung infections in intense care wards,
[43] so-called
acute abdomen,
[44] middle ear infections, sinusitis and throat infections,
[45] and penetrating gut injuries.
[46][47] In some situations a short course may not cure the infection as well as a long course.
[48] A
BMJ editorial recommended that antibiotics can often be safely stopped 72 hours after symptoms resolve.
[49]
Because individuals may feel better before the infection is
eradicated, doctors must provide instructions to them so they know when
it is safe to stop taking a prescription. Some researchers advocate
doctors' using a very short course of antibiotics, reevaluating the
patient after a few days, and stopping treatment if there are no
clinical signs of infection.
[50]
Certain
antibiotic classes result in resistance more than others. Increased rates of MRSA infections are seen when using
glycopeptides,
cephalosporins, and
quinolone antibiotics.
[51][52] Cephalosporins, and particularly quinolones and
clindamycin, are more likely to produce colonisation with
Clostridium difficile.
[53][54]
Factors within the intensive care unit setting such as mechanical
ventilation and multiple underlying diseases also appear to contribute
to bacterial resistance.
[55] Poor hand
hygiene by hospital staff has been associated with the spread of resistant organisms.
[56]
Veterinary medicine
All
animals carry bacteria in their intestines. Antibiotics are given to
animals. Antibiotics kill most bacteria. But resistant bacteria survive
and multiply.
The
World Health Organization
concluded that inappropriate use of antibiotics in animal husbandry is
an underlying contributor to the emergence and spread of
antibiotic-resistant germs, and that the use of antibiotics as growth
promoters in animal feeds should be restricted.
[57] The
World Organisation for Animal Health
has added to the Terrestrial Animal Health Code a series of guidelines
with recommendations to its members for the creation and harmonization
of national antimicrobial resistance surveillance and monitoring
programs,
[58] monitoring of the quantities of antibiotics used in animal husbandry,
[59]
and recommendations to ensure the proper and prudent use of antibiotic
substances. Another guideline is to implement methodologies that help to
establish associated risk factors and assess the risk of antibiotic
resistance.
[60]
Natural occurrence
Naturally occurring antibiotic resistance is common.
[61] Genes for resistance to antibiotics, like antibiotics themselves, are ancient.
[62][63] The genes that confer resistance are known as the environmental
resistome.
[61]
These genes may be transferred from non-disease-causing bacteria to
those that do cause disease, leading to clinically significant
antibiotic resistance.
[61]
In 1952 it was shown that penicillin-resistant bacteria existed before penicillin treatment;
[64] and also preexistent bacterial resistance to
streptomycin.
[65] In 1962, the presence of
penicillinase was detected in dormant
endospores of
Bacillus licheniformis, revived from dried soil on the roots of plants, preserved since 1689 in the
British Museum.
[66][67][68] Six
strains of
Clostridium, found in the bowels of William Braine and John Hartnell (members of the
Franklin Expedition) showed resistance to
cefoxitin and
clindamycin.
[69]
Penicillinase may have emerged as a defense mechanism for bacteria in their
habitats, such as the case of penicillinase-rich
Staphylococcus aureus, living with penicillin-producing
Trichophyton; however, this may be circumstantial.
[68] Search for a penicillinase ancestor has focused on the class of
proteins that must be
a priori capable of specific combination with
penicillin.
[70]
The resistance to cefoxitin and clindamycin in turn was attributed to
Braine's and Hartnell's contact with microorganisms that naturally
produce them or
random mutation in the
chromosomes of
Clostridium strains.
[69]
There is evidence that
heavy metals and other pollutants may select for antibiotic-resistant bacteria, generating a constant source of them in small numbers.
[71]
Water pollution
Antibiotic
resistance is a growing problem among humans and wildlife in
terrestrial or aquatic environments. In this respect, the spread and
contamination of the environment, especially through
water pollution "hot spots" such as hospital
wastewater and untreated urban wastewater, is a growing and serious public health problem.
[74][75] Antibiotics have been polluting the environment since their introduction through
human waste (medication, farming), animals, and the pharmaceutical industry.
[76]
The contribution of the pharmaceutical industry is so significant that
parallels can be drawn between countries with highest rate of increasing
antibiotic resistance and countries with largest footprint of
pharmaceutical industry. China, which contributes to nearly 90 per cent
of the world's active pharmaceutical ingredient (API) manufacturing, has
seen a 22 per cent increase in rate of antimicrobial resistance in six
years, compared to a 6 per cent increase in the United States.
[77]
Along with antibiotic waste, resistant bacteria follow, thus
introducing antibiotic-resistant bacteria into the environment. Already
in 2011, mapping of sewage and water supply samples in
New Delhi showed widespread and uncontrolled infection as indicated by the presence of NDM-1-positive enteric bacteria (
New Delhi metallo-beta-lactamase 1).
[78]
As bacteria replicate quickly, the resistant bacteria that enter
water bodies through wastewater replicate their resistance genes as they
continue to divide. In addition, bacteria carrying resistance genes
have the ability to spread those genes to other species via horizontal
gene transfer. Therefore, even if the specific antibiotic is no longer
introduced into the environment, antibiotic-resistance genes will
persist through the bacteria that have since replicated without
continuous exposure.
[76]
Antibiotic resistance is widespread in marine vertebrates, and they may
be important reservoirs of antibiotic-resistant bacteria in the
marine environment.
[79]
Prevention
Mission Critical: Preventing Antibiotic Resistance (CDC report, 2014)
There have been increasing public calls for global collective action
to address the threat, including a proposal for international treaty on
antimicrobial resistance. Further detail and attention is still needed
in order to recognize and measure trends in resistance on the
international level; the idea of a global tracking system has been
suggested but implementation has yet to occur. A system of this nature
would provide insight to areas of high resistance as well as information
necessary for evaluation of programs and other changes made to fight or
reverse antibiotic resistance.
Five important strategies needed for minimising antibiotic resistance are as follows:
[80]
- Antibiotic stewardship to maintain the value of existing and future antibiotics
- The timing of prescription to use the effective antibiotics sooner rather than later
- To develop and approve ten new antibiotics by 2020
- Development of a molecular method for detecting antibiotic resistance genes
- To avoid the delay in distribution of US$2 billion global antibiotic resistance innovation fund.
Duration of antibiotics
Antibiotic treatment duration should be based on the infection and other health problems a person may have.
[10] For many infections once a person has improved there is little evidence that stopping treatment causes more resistance.
[10] Some therefore feel that stopping early may be reasonable in some cases.
[10] Other infections, however, do require long courses regardless of whether a person feels better.
[10]
Monitoring and mapping
There are multiple national and international monitoring programs for drug-resistant threats, including
methicillin-resistant Staphylococcus aureus (MRSA),
vancomycin-resistant S. aureus (VRSA),
extended spectrum beta-lactamase (ESBL),
vancomycin-resistant Enterococcus (VRE),
multidrug-resistant A. baumannii (MRAB).
[81]
ResistanceOpen is an online global map of antimicrobial resistance developed by
HealthMap which displays aggregated data on antimicrobial resistance from publicly available and user submitted data.
[82][83] The website can display data for a 25-mile radius from a location. Users may submit data from
antibiograms
for individual hospitals or laboratories. European data is from the
EARS-Net (European Antimicrobial Resistance Surveillance Network), part
of the
ECDC.
ResistanceMap is a website by the
Center for Disease Dynamics, Economics & Policy and provides data on antimicrobial resistance on a global level.
[84]
Limiting antibiotic use
Antibiotic stewardship programmes appear useful in reducing rates of antibiotic resistance.
[85]
Excessive antibiotic use has become one of the top contributors
to the development of antibiotic resistance. Since the beginning of the
antibiotic era, antibiotics have been used to treat a wide range of
disease.
[86]
Overuse of antibiotics has become the primary cause of rising levels of
antibiotic resistance. The main problem is that doctors are willing to
prescribe antibiotics to ill-informed individuals who believe that
antibiotics can cure nearly all illnesses, including viral infections
like the common cold. In an analysis of drug prescriptions, 36% of
individuals with a cold or an upper respiratory infection (both viral in
origin) were given prescriptions for antibiotics.
[87] These prescriptions accomplished nothing other than increasing the risk of further evolution of antibiotic resistant bacteria.
At the hospital level
Antimicrobial stewardship teams in hospitals are encouraging optimal use of antimicrobials.
[88]
The goals of antimicrobial stewardship are to help practitioners pick
the right drug at the right dose and duration of therapy while
preventing misuse and minimizing the development of resistance. Stewardship may reduce the length of stay by an average of slightly over
1 day while not increasing the risk of death.
[89]
At the level of GP
Given
the volume of care provided in primary care (General Practice), recent
strategies have focused on reducing unnecessary antibiotic prescribing
in this setting. Simple interventions, such as written information
explaining the futility of antibiotics for common infections such as
upper respiratory tract infections, have been shown to reduce antibiotic
prescribing.
[90]
The prescriber should closely adhere to the five rights of drug
administration: the right patient, the right drug, the right dose, the
right route, and the right time.
[91]
Cultures should be taken before treatment when indicated and treatment potentially changed based on the susceptibility report.
[11][92]
About a third of antibiotic prescriptions written in
outpatient settings
in the United States were not appropriate in 2010 and 2011. Doctors in
the U.S. wrote 506 annual antibiotic scripts for every 1,000 people,
with 353 being medically necessary.
[93]
Health workers and pharmacists can help tackle resistance by:
enhancing infection prevention and control; only prescribing and
dispensing antibiotics when they are truly needed; prescribing and
dispensing the right antibiotic(s) to treat the illness.
[22]
At the individual level
People
can help tackle resistance by using antibiotics only when prescribed by
a doctor; completing the full prescription, even if they feel better;
never sharing antibiotics with others or using leftover prescriptions.
[22]
Country examples
- The Netherlands has the lowest rate of antibiotic prescribing in the OECD, at a rate of 11.4 defined daily doses (DDD) per 1,000 people per day in 2011.
- Germany and Sweden also have lower prescribing rates, with Sweden's rate having been declining since 2007.
- Greece, France and Belgium have high prescribing rates of more than 28 DDD.[94]
Water, sanitation, hygiene
Infectious
disease control through improved water, sanitation and hygiene (WASH)
infrastructure needs to be placed at the center of the antimicrobial
resistance (AMR) agenda. The spread of infectious diseases caused by
inadequate WASH standards is a major driver of antibiotic demand in
developing countries.
[35]
Growing usage of antibiotics together with persistent infectious
disease levels have led to a dangerous cycle in which reliance on
antimicrobials increases while the efficacy of drugs diminishes.
[35] The proper use of infrastructure for
water, sanitation and hygiene (WASH)
can result in a 47–72 percent decrease of diarrhea cases treated with
antibiotics depending on the type of intervention and its effectiveness.
[35]
A reduction of the diarrhea disease burden through improved
infrastructure would result in large decreases in the number of diarrhea
cases treated with antibiotics. This was estimated as ranging from 5
million in Brazil to up to 590 million in India by the year 2030.
[35]
The strong link between increased consumption and resistance indicates
that this will directly mitigate the accelerating spread of AMR.
[35] Sanitation and water for all by 2030 is
Goal Number 6 of the
Sustainable Development Goals.
An increase in
hand washing compliance by hospital staff results in decreased rates of resistant organisms.
[95]
Management in animal use
Europe
In 1997, European Union health ministers voted to ban
avoparcin and four additional antibiotics used to promote animal growth in 1999.
[96]
In 2006 a ban on the use of antibiotics in European feed, with the
exception of two antibiotics in poultry feeds, became effective.
[97] In Scandinavia, there is evidence that the ban has led to a lower
prevalence of antibiotic resistance in (nonhazardous) animal bacterial populations.
[98]
As of 2004, several European countries established a decline of
antimicrobial resistance in humans through limiting the usage
antimicrobials in agriculture and food industries without jeopardizing
animal health or economic cost.
[99]
United States
The
United States Department of Agriculture (USDA) and the
Food and Drug Administration (FDA) collect data on antibiotic use in humans and in a more limited fashion in animals.
[100]
The FDA first determined in 1977 that there is evidence of emergence of
antibiotic-resistant bacterial strains in livestock. The
long-established practice of permitting OTC sales of antibiotics
(including penicillin and other drugs) to lay animal owners for
administration to their own animals nonetheless continued in all states.
In 2000, the FDA announced their intention to revoke approval of
fluoroquinolone use in poultry production because of substantial evidence linking it to the emergence of fluoroquinolone-resistant
Campylobacter
infections in humans. Legal challenges from the food animal and
pharmaceutical industries delayed the final decision to do so until
2006.
[101]
Fluroquinolones have been banned from extra-label use in food animals
in the USA since 2007. However, they remain widely used in companion and
exotic animals.
Global action plans and awareness
The
increasing interconnectedness of the world and the fact that new
classes of antibiotics have not been developed and approved for more
than 25 years highlight the extent to which antimicrobial resistance is a
global health challenge.
[102]
A global action plan to tackle the growing problem of resistance to
antibiotics and other antimicrobial medicines was endorsed at the
Sixty-eighth World Health Assembly in May 2015.
[103]
One of the key objectives of the plan is to improve awareness and
understanding of antimicrobial resistance through effective
communication, education and training. This global action plan developed
by the World Health Organization was created to combat the issue of
antimicrobial resistance and was guided by the advice of countries and
key stakeholders. The WHO's global action plan is composed of five key
objectives that can be targeted through different means, and represents
countries coming together to solve a major problem that can have future
health consequences.
[103]
- React based in Sweden has produced informative material on AMR for the general public.[104]
- Videos are being produced for the general public to generate interest and awareness.[105][106]
Antibiotic Awareness Week
The
World Health Organization has promoted the first World Antibiotic
Awareness Week running from 16–22 November 2015. The aim of the week is
to increase global awareness of antibiotic resistance. It also wants to
promote the correct usage of antibiotics across all fields in order to
prevent further instances of antibiotic resistance.
[107]
World Antibiotic Awareness Week has been held every November
since 2015. For 2017, the Food and Agriculture Organization of the
United Nations (FAO), the World Health Organization (WHO) and the World
Organisation for Animal Health (OIE) are together calling for
responsible use of antibiotics in humans and animals to reduce the
emergence of antibiotic resistance.
[108]
Mechanisms and organisms
Fundamentals
Diagram
depicting antibiotic resistance through alteration of the antibiotic's
target site, modeled after MRSA's resistance to penicillin. Beta-lactam
antibiotics permanently inactivate
PBP enzymes, which are essential for bacterial life, by permanently binding to their active sites.
MRSA, however, expresses a PBP that does not allow the antibiotic into its active site.
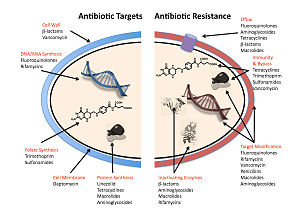
The four main mechanisms by which microorganisms exhibit resistance to antimicrobials are:
- Drug inactivation or modification: for example, enzymatic deactivation of penicillin G in some penicillin-resistant bacteria through the production of β-lactamases. The emergence of carbapenem-resistant Gram-negative pathogens poses a serious threat to public health worldwide. Klebsiella pneumoniae
carbapenemases (KPCs) and carbapenemases of the oxacillinase-48
(OXA-48) type have been reported worldwide. New Delhi
metallo-β-lactamase (NDM) carbapenemases were originally identified in
Sweden in 2008 and have spread worldwide rapidly.[109]
Most commonly, the protective enzymes produced by the bacterial cell
will add an acetyl or phosphate group to a specific site on the
antibiotic, which will reduce its ability to bind to the bacterial
ribosomes and disrupt protein synthesis.[110]
- Alteration of target- or binding site: for example, alteration of PBP—the binding target site of penicillins—in MRSA
and other penicillin-resistant bacteria. Another protective mechanism
found among bacterial species is ribosomal protection proteins. These
proteins protect the bacterial cell from antibiotics that target the
cell’s ribosomes to inhibit protein synthesis. The mechanism involves
the binding of the ribosomal protection proteins to the ribosomes of the
bacterial cell, which in turn changes its conformational shape. This
allows the ribosomes to continue synthesizing proteins essential to the
cell while preventing antibiotics from binding to the ribosome to
inhibit protein synthesis.[111]
- Alteration of metabolic pathway: for example, some sulfonamide-resistant bacteria do not require para-aminobenzoic acid (PABA), an important precursor for the synthesis of folic acid and nucleic acids in bacteria inhibited by sulfonamides, instead, like mammalian cells, they turn to using preformed folic acid.[112]
- Reduced drug accumulation: by decreasing drug permeability or increasing active efflux (pumping out) of the drugs across the cell surface[113]
These pumps within the cellular membrane of certain bacterial species
are used to pump antibiotics out of the cell before they are able to do
any damage. They are often activated by a specific substrate associated
with an antibiotic.[114] as in fluoroquinolone resistance.[115]
A number of mechanisms used by common antibiotics to deal with bacteria and ways by which bacteria become resistant to them.
Antibiotic resistance can be a result of
horizontal gene transfer,
[116] and also of unlinked point mutations in the
pathogen genome at a rate of about 1 in 10
8
per chromosomal replication. Mutations are rare but the fact that
bacteria reproduce at such a high rate allows for the effect to be
significant. A mutation may produce a change in the binding site of the
antibiotic, which may allow the site to continue proper functioning in
the presence of the antibiotic or prevent the binding of the antibiotic
to the site altogether.
[117]
Antibiotic action against a pathogen can be seen as an
environmental pressure. Those bacteria with a mutation that allows them
to survive will reproduce, pass the trait to their offspring, which
leads to the
microevolution
of a fully resistant colony. Chromosomal mutations providing antibiotic
resistance benefit the bacteria but also confer a cost of fitness. For
example, a ribosomal mutation may protect a bacterial cell by changing
the binding site of an antibiotic but will also slow protein synthesis.
[110] manifesting, in slower growth rate.
[118]
In Gram-negative bacteria, plasmid-mediated resistance genes produce proteins that can bind to
DNA gyrase, protecting it from the action of quinolones. Finally, mutations at key sites in DNA gyrase or
topoisomerase IV can decrease their binding affinity to quinolones, decreasing the drug's effectiveness.
[119]
Bacteria
Bacteria can often develop
antibiotic resistance.
Mutations that confer increased survival are selected for in
natural selection,
which can happen quickly in bacteria because lifespans and production
of new generations can be on a timescale of mere hours. A new (de novo)
mutation in a parent cell can quickly become an
inherited
mutation of widespread prevalence. Moreover, some adaptive mutations
can propagate not only through inheritance but also through
horizontal gene transfer via
plasmids, followed by inheritance from those parents.
Recent findings show no necessity of large populations of
bacteria for the appearance of antibiotic resistance. Small populations
of
E. coli in an antibiotic gradient can become resistant. Any
heterogeneous environment with respect to nutrient and antibiotic
gradients may facilitate antibiotic resistance in small bacterial
populations. Researchers hypothesize that the mechanism of resistance
development is based on four SNP mutations in the genome of
E. coli produced by the gradient of antibiotic.
[120]
Antibiotic resistance can be introduced artificially into a microorganism through laboratory protocols, sometimes used as a
selectable marker
to examine the mechanisms of gene transfer or to identify individuals
that absorbed a piece of DNA that included the resistance gene and
another gene of interest.
[121]
New Delhi metallo-beta-lactamase 1 (NDM-1)
[122] is an
enzyme that makes
bacteria resistant to a broad range of
beta-lactam antibiotics. The most common bacteria that make this enzyme are
gram-negative such as
Escherichia coli and
Klebsiella pneumoniae, but the gene for NDM-1 can spread from one strain of bacteria to another by
horizontal gene transfer.
[123]
Viruses
Specific
antiviral drugs
are used to treat some viral infections. These drugs prevent viruses
from reproducing by inhibiting essential stages of the virus's
replication cycle in infected cells. Antivirals are used to treat
HIV,
hepatitis B,
hepatitis C,
influenza,
herpes viruses including
varicella zoster virus,
cytomegalovirus and
Epstein-Barr virus. With each virus, some strains have become resistant to the administered drugs.
[124]
Resistance to HIV antivirals is problematic, and even multi-drug resistant strains have evolved.
[125] Resistant strains of the HIV virus emerge rapidly if only one antiviral drug is used.
[126]
Using three or more drugs together has helped to control this problem,
but new drugs are needed because of the continuing emergence of
drug-resistant HIV strains.
[127]
Fungi
Infections by fungi are a cause of high morbidity and mortality in
immunocompromised persons, such as those with HIV/AIDS, tuberculosis or receiving
chemotherapy.
[128] The fungi
candida,
Cryptococcus neoformans and
Aspergillus fumigatus cause most of these infections and antifungal resistance occurs in all of them.
[129]
Multidrug resistance in fungi is increasing because of the widespread
use of antifungal drugs to treat infections in immunocompromised
individuals.
[130]
Of particular note,
Fluconazole-resistant Candida species have been highlighted as a growing problem by the CDC.
[81] More than 20 species of Candida can cause
Candidiasis infection, the most common of which is
Candida albicans.
Candida yeasts normally inhabit the skin and mucous membranes without
causing infection. However, overgrowth of Candida can lead to
Candidiasis. Some Candida strains are becoming resistant to first-line
and second-line
antifungal agents such as
azoles and
echinocandins.
[81]
Parasites
The
protozoan parasites that cause the diseases
malaria,
trypanosomiasis,
toxoplasmosis,
cryptosporidiosis and
leishmaniasis are important human pathogens.
[131]
Malarial parasites that are resistant to the drugs that are
currently available to infections are common and this has led to
increased efforts to develop new drugs.
[132] Resistance to recently developed drugs such as
artemisinin has also been reported. The problem of drug resistance in malaria has driven efforts to develop vaccines.
[133]
Trypanosomes are parasitic protozoa that cause
African trypanosomiasis and
Chagas disease (American trypanosomiasis).
[134][135] There are no vaccines to prevent these infections so drugs such as
pentamidine and
suramin,
benznidazole and
nifurtimox are used to treat infections. These drugs are effective but infections caused by resistant parasites have been reported.
[131]
Leishmaniasis
is caused by protozoa and is an important public health problem
worldwide, especially in sub-tropical and tropical countries. Drug
resistance has "become a major concern".
[136]
History
The
discovery of penicillin in 1928 and other antibiotics in the 20th
century proved to be a significant medical achievement, saving millions
of lives and significantly reducing the burden of infectious diseases.
[137]
The 1950s to 1970s represented the golden age of antibiotic discovery,
where countless new classes of antibiotics were discovered to treat
previously incurable diseases such as tuberculosis and syphilis.
[138]
However, since that time the discovery of new classes of antibiotics
has been almost nonexistent, and represents a situation that is
especially problematic considering the resiliency of bacteria shown over
time and the continued misuse and overuse of antibiotics in treatment.
[139]
The phenomenon of antimicrobial resistance caused by overuse of antibiotics was predicted by
Alexander Fleming
who said "The time may come when penicillin can be bought by anyone in
the shops. Then there is the danger that the ignorant man may easily
under-dose himself and by exposing his microbes to nonlethal quantities
of the drug make them resistant."
[140][141]
Without the creation of new and stronger antibiotics an era where
common infections and minor injuries can kill, and where complex
procedures such as surgery and chemotherapy become too risky, is a very
real possibility.
[142]
Antimicrobial resistance threatens the world as we know it, and can
lead to epidemics of enormous proportions if preventive actions are not
taken. In this day and age current antimicrobial resistance leads to
longer hospital stays, higher medical costs, and increased mortality.
[139]
Society and culture
For the
fiscal year
2016 budget, President Obama suggested to nearly double the amount of
federal funding to "combat and prevent" antibiotic resistance to more
than $1.2 billion.
[143] Many international funding agencies like USAID, DFID,
SIDA and
Bill & Melinda Gates Foundation have pledged money for developing strategies to counter antimicrobial resistance.
Since the mid-1980s pharmaceutical companies have invested in
medications for cancer or chronic disease that have greater potential to
make money and have "de-emphasized or dropped development of
antibiotics".
[144] On January 20, 2016 at the
World Economic Forum in
Davos,
Switzerland,
more than "80 pharmaceutical and diagnostic companies" from around the
world called for "transformational commercial models" at a global level
to spur research and development on antibiotics and on the "enhanced use
of diagnostic tests that can rapidly identify the infecting organism".
[144]
Legal frameworks
Some
global health scholars have argued that a global, legal framework is
needed to prevent and control antimicrobial resistance.
[145][146][19][147]
For instance, binding global policies could be used to create
antimicrobial use standards, regulate antibiotic marketing, and
strengthen global surveillance systems.
[19][145] Ensuring compliance of involved parties is a challenge.
[19]
Global antimicrobial resistance policies could take lessons from the
environmental sector by adopting strategies that have made international
environmental agreements successful in the past such as: sanctions for
non-compliance, assistance for implementation, majority vote
decision-making rules, an independent scientific panel, and specific
commitments.
[148]
U.S.
On March 27, 2015, the
White House
released a comprehensive plan to address the increasing need for
agencies to combat the rise of antibiotic-resistant bacteria. The Task
Force for Combating Antibiotic-Resistant Bacteria developed
The National Action Plan for Combating Antibiotic-Resistant Bacteria
with the intent of providing a roadmap to guide the US in the
antibiotic resistance challenge and with hopes of saving many lives.
This plan outlines steps taken by the Federal government over the next
five years needed in order to prevent and contain outbreaks of
antibiotic-resistant infections; maintain the efficacy of antibiotics
already on the market; and to help to develop future diagnostics,
antibiotics, and vaccines.
[149]
The Action Plan was developed around five goals with focuses on
strengthening health care, public health veterinary medicine,
agriculture, food safety and research, and manufacturing. These goals,
as listed by the White House, are as follows:
- Slow the Emergence of Resistant Bacteria and Prevent the Spread of Resistant Infections
- Strengthen National One-Health Surveillance Efforts to Combat Resistance
- Advance Development and use of Rapid and Innovative Diagnostic Tests
for Identification and Characterization of Resistant Bacteria
- Accelerate Basic and Applied Research and Development for New Antibiotics, Other Therapeutics, and Vaccines
- Improve International Collaboration and Capacities for Antibiotic
Resistance Prevention, Surveillance, Control and Antibiotic Research and
Development
The following are goals set to meet by 2020:
[149]
- Establishment of antimicrobial programs within acute care hospital settings
- Reduction of inappropriate antibiotic prescription and use by at least 50% in outpatient settings and 20% inpatient settings
- Establishment of State Antibiotic Resistance (AR) Prevention Programs in all 50 states
- Elimination of the use of medically important antibiotics for growth promotion in food-producing animals.
Policies
According to
WHO
policymakers can help tackle resistance by strengthening resistance
tracking and laboratory capacity; regulating and promoting appropriate
use of medicines.
[22]
Policymakers and industry can help tackle resistance by: fostering
innovation and research and development of new tools; promoting
cooperation and information sharing among all stakeholders.
[22]
Further research
It is unclear if rapid viral testing affects antibiotic use in children.
[150]
Vaccines
Microorganisms do not develop resistance to
vaccines
because a vaccine enhances the body's immune system, whereas an
antibiotic operates separately from the body's normal defenses.
Furthermore, if the use of vaccines increase, there is evidence that
antibiotic resistant strains of pathogens will decrease; the need for
antibiotics will naturally decrease as vaccines prevent infection before
it occurs.
[151] However, new strains that escape immunity induced by vaccines may
evolve; for example, an updated
influenza vaccine is needed each year.
While theoretically promising, antistaphylococcal vaccines have
shown limited efficacy, because of immunological variation between
Staphylococcus
species, and the limited duration of effectiveness of the antibodies
produced. Development and testing of more effective vaccines is
underway.
[152]
Alternating therapy
Alternating
therapy is a proposed method in which two or three antibiotics are
taken in a rotation versus taking just one antibiotic such that bacteria
resistant to one antibiotic are killed when the next antibiotic is
taken. Studies have found that this method reduces the rate at which
antibiotic resistant bacteria emerge in vitro relative to a single drug
for the entire duration.
[153]
Studies have found that bacteria that evolve antibiotic
resistance towards one group of antibiotic may become more sensitive
others.
[154] This phenomona can be utilized to select against resistant bacteria using an approach termed collateral sensitivity cycling,
[155] which has recently been found to be relevant in developing treatment strategies for chronic infections caused by
Pseudomonas aeruginosa.
[156]
Development of new drugs
Since the discovery of antibiotics,
research and development
(R&D) efforts have provided new drugs in time to treat bacteria
that became resistant to older antibiotics, but in the 2000s there has
been concern that development has slowed enough that seriously ill
people may run out of treatment options.
[157]
Another concern is that doctors may become reluctant to perform routine
surgeries because of the increased risk of harmful infection.
[158] Backup treatments can have serious side-effects; for example, treatment of
multi-drug-resistant tuberculosis can cause deafness or psychological disability.
[159] The potential crisis at hand is the result of a marked decrease in industry R&D.
[160] Poor financial investment in antibiotic research has exacerbated the situation.
[161][160] The pharmaceutical industry has little incentive to invest in
antibiotics because of the high risk and because the potential financial
returns are less likely to cover the cost of
development than for other pharmaceuticals.
[162] In 2011,
Pfizer,
one of the last major pharmaceutical companies developing new
antibiotics, shut down its primary research effort, citing poor
shareholder returns relative to drugs for chronic illnesses.
[163] However, small and medium-sized pharmaceutical companies are still active in antibiotic drug research.
In the United States, drug companies and the administration of President
Barack Obama have been proposing changing the standards by which the FDA approves antibiotics targeted at resistant organisms.
[158][164] On 12 December 2013, the Antibiotic Development to Advance Patient Treatment (ADAPT) Act of 2013 was introduced in the
U.S. Congress.
The ADAPT Act aims to fast-track the drug development in order to
combat the growing public health threat of 'superbugs'. Under this Act,
the FDA can approve antibiotics and antifungals needed for
life-threatening infections based on data from smaller clinical trials.
The
Centers for Disease Control and Prevention
(CDC) will reinforce the monitoring of the use of antibiotics that
treat serious and life-threatening infections and the emerging
resistance, and make the data publicly available. The FDA antibiotics
labeling process, 'Susceptibility Test Interpretive Criteria for
Microbial Organisms' or 'breakpoints' is also streamlined to allow the
most up-to-date and cutting-edge data available to healthcare
professionals under the new Act.
[165][166]
On 18 September 2014 Obama signed an executive order
[167] to implement the recommendations proposed in a report
[168] by the
President's Council of Advisors on Science and Technology
(PCAST) which outlines strategies to stream-line clinical trials and
speed up the R&D of new antibiotics. Among the proposals:
- Create a 'robust, standing national clinical trials network for
antibiotic testing' which will promptly enroll patients once identified
to be suffering from dangerous bacterial infections. The network will
allow testing multiple new agents from different companies
simultaneously for their safety and efficacy.
- Establish a 'Special Medical Use (SMU)' pathway for FDA to approve
new antimicrobial agents for use in limited patient populations, shorten
the approval timeline for new drug so patients with severe infections
could benefit as quickly as possible.
- Provide economic incentives, especially for development of new
classes of antibiotics, to offset the steep R&D costs which drive
away the industry to develop antibiotics.
The executive order also included a $20 million prize to encourage
the development of diagnostic tests to identify highly resistant
bacterial infections.
[169]
The U.S.
National Institutes of Health plans to fund a new research network on the issue up to $62 million from 2013 to 2019.
[170] Using authority created by the
Pandemic and All Hazards Preparedness Act of 2006, the
Biomedical Advanced Research and Development Authority in the U.S.
Department of Health and Human Services
announced that it will spend between $40 million and $200 million in
funding for R&D on new antibiotic drugs under development by
GlaxoSmithKline.
[171]
Phage therapy
Phage therapy is the
therapeutic use of
bacteriophages to treat
pathogenic bacterial infections.
[172] Phage therapy has many potential applications in human medicine as well as dentistry, veterinary science, and agriculture.
[173]
Phage therapy relies on the use of naturally-occurring
bacteriophages to infect and lyse bacteria at the site of infection in a
host. Due to current advances in genetics and biotechnology these
bacteriophages can possibly be manufactured to treat specific
infections.
[174]
Phages can be bioengineered to target multidrug-resistant bacterial
infections, and their use involves the added benefit of preventing the
elimination of beneficial bacteria in the human body.
[174]
Phages destroy bacterial cell walls and membrane through the use of
lytic proteins which kill bacteria by making many holes from the inside
out.
[175]
Bacteriophages can even possess the ability to digest the biofilm that
many bacteria develop that protect them from antibiotics in order to
effectively infect and kill bacteria. Bioengineering can play a role in
creating successful bacteriophages.
[175]
Bacteriophages are much more specific than
antibiotics. They are typically harmless not only to the host
organism, but also to other beneficial bacteria, such as the
gut flora, reducing the chances of
opportunistic infections.
[176]
Bacteriophages are used against antibiotic resistant bacteria in
Georgia (
George Eliava Institute) and in one institute in
Wrocław, Poland.