| Human mitochondrial DNA | |
|---|---|

The 16,569 bp long human mitochondrial genome with the protein-coding, ribosomal RNA, and transfer RNA genes.
| |
| Features | |
| Length (bp) | 16,569 |
| No. of genes | 13 (coding genes) 24 (non coding genes) |
| Type | Mitochondrial DNA |
| Complete gene lists | |
| HGNC | Gene list |
| NCBI | Gene list |
| External map viewers | |
| Ensembl | Chromosome MT |
| Entrez | Chromosome MT |
| NCBI | Chromosome MT |
| UCSC | Chromosome M |
| Full DNA sequences | |
| RefSeq | NC_012920 (FASTA) |
| GenBank | J01415 (FASTA) |
Human mitochondrial genetics is the study of the genetics of human mitochondrial DNA (the DNA contained in human mitochondria). The human mitochondrial genome is the entirety of hereditary information contained in human mitochondria. Mitochondria are small structures in cells that generate energy for the cell to use, and are hence referred to as the "powerhouses" of the cell.
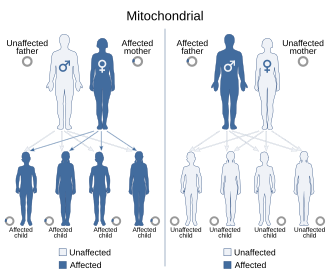
Mitochondrial DNA (mtDNA) is not transmitted through nuclear DNA (nDNA). In humans, as in most multicellular organisms, mitochondrial DNA is inherited only from the mother's ovum. There are theories, however, that paternal mtDNA transmission in humans can occur under certain circumstances.
Mitochondrial inheritance is therefore non-Mendelian, as Mendelian inheritance presumes that half the genetic material of a fertilized egg (zygote) derives from each parent.
Eighty percent of mitochondrial DNA codes for mitochondrial RNA, and therefore most mitochondrial DNA mutations lead to functional problems, which may be manifested as muscle disorders (myopathies).
Because they provide 30 molecules of ATP per glucose molecule in contrast to the 2 ATP molecules produced by glycolysis, mitochondria are essential to all higher organisms for sustaining life. The mitochondrial diseases are genetic disorders carried in mitochondrial DNA, or nuclear DNA coding for mitochondrial components. Slight problems with any one of the numerous enzymes used by the mitochondria can be devastating to the cell, and in turn, to the organism.
Quantity
In humans, mitochondrial DNA (mtDNA) forms closed circular molecules that contain 16,569 DNA base pairs,
with each such molecule normally containing a full set of the
mitochondrial genes. Each human mitochondrion contains, on average,
approximately 5 such mtDNA molecules, with the quantity ranging between 1
and 15. Each human cell contains approximately 100 mitochondria, giving a total number of mtDNA molecules per human cell of approximately 500.
Inheritance patterns
Mitochondrial inheritance patterns
The
reason for maternal inheritance in mitochondrial DNA is that when the
sperm enters the egg cell, it discards its middle part, which contains
its mitochondria, so that only its head with the nucleus penetrates the
egg cell.
Because mitochondrial diseases
(diseases due to malfunction of mitochondria) can be inherited both
maternally and through chromosomal inheritance, the way in which they
are passed on from generation to generation can vary greatly depending
on the disease. Mitochondrial genetic mutations that occur in the
nuclear DNA can occur in any of the chromosomes (depending on the
species). Mutations inherited through the chromosomes can be autosomal
dominant or recessive and can also be sex-linked dominant or recessive.
Chromosomal inheritance follows normal Mendelian laws, despite the fact that the phenotype of the disease may be masked.
Because of the complex ways in which mitochondrial and nuclear
DNA "communicate" and interact, even seemingly simple inheritance is
hard to diagnose. A mutation in chromosomal DNA may change a protein
that regulates (increases or decreases) the production of another
certain protein in the mitochondria or the cytoplasm; this may lead to
slight, if any, noticeable symptoms. On the other hand, some devastating
mtDNA mutations are easy to diagnose because of their widespread damage
to muscular, neural, and/or hepatic tissues (among other high-energy
and metabolism-dependent tissues) and because they are present in the
mother and all the offspring.
The number of affected mtDNA molecules inherited by a specific offspring can vary greatly because
- the mitochondria within the fertilized oocyte is what the new life will have to begin with (in terms of mtDNA),
- the number of affected mitochondria varies from cell (in this case, the fertilized oocyte) to cell depending both on the number it inherited from its mother cell and environmental factors which may favor mutant or wildtype mitochondrial DNA,
- the number of mtDNA molecules in the mitochondria varies from around two to ten.
It is possible, even in twin births, for one baby to receive more
than half mutant mtDNA molecules while the other twin may receive only a
tiny fraction of mutant mtDNA molecules with respect to wildtype
(depending on how the twins divide from each other and how many mutant
mitochondria happen to be on each side of the division). In a few cases,
some mitochondria or a mitochondrion from the sperm cell enters the
oocyte but paternal mitochondria are actively decomposed.
Genes
Electron transport chain, and humanin
It
was originally incorrectly believed that the mitochondrial genome
contained only 13 protein-coding genes, all of them encoding proteins of
the electron transport chain. However, in 2001, a 14th biologically active protein called humanin was discovered, and was found to be encoded by the mitochondrial gene MT-RNR2 which also encodes part of the mitochondrial ribosome (made out of RNA):
| Complex number |
Category | Genes | Positions in the mitogenome | Strand |
|---|---|---|---|---|
| I | NADH dehydrogenase | |||
| MT-ND1 | 3,307–4,262 | L | ||
| MT-ND2 | 4,470–5,511 | L | ||
| MT-ND3 | 10,059–10,404 | L | ||
| MT-ND4L | 10,470–10,766 | L | ||
| MT-ND4 | 10,760–12,137 (overlap with MT-ND4L) | L | ||
| MT-ND5 | 12,337–14,148 | L | ||
| MT-ND6 | 14,149–14,673 | H | ||
| III | Coenzyme Q - cytochrome c reductase / Cytochrome b | MT-CYB | 14,747–15,887 | L |
| IV | Cytochrome c oxidase | MT-CO1 | 5,904–7,445 | L |
| MT-CO2 | 7,586–8,269 | L | ||
| MT-CO3 | 9,207–9,990 | L | ||
| V | ATP synthase | MT-ATP6 | 8,527–9,207 (overlap with MT-ATP8) | L |
| MT-ATP8 | 8,366–8,572 | L | ||
| — | Humanin | MT-RNR2 | — | — |
Unlike the other proteins, humanin does not remain in the
mitochondria, and interacts with the rest of the cell and cellular
receptors. Humanin can protect brain cells by inhibiting apoptosis. Despite its name, versions of humanin also exist in other animals, such as rattin in rats.
rRNA
The following genes encode rRNAs:
| Subunit | rRNA | Genes | Positions in the mitogenome | Strand |
|---|---|---|---|---|
| Small (SSU) | 12S | MT-RNR1 | 648–1,601 | L |
| Large (LSU) | 16S | MT-RNR2 | 1,671–3,229 | L |
tRNA
The following genes encode tRNAs:
| Amino Acid | 3-Letter | 1-Letter | MT DNA | Positions | Strand |
|---|---|---|---|---|---|
| Alanine | Ala | A | MT-TA | 5,587–5,655 | H |
| Arginine | Arg | R | MT-TR | 10,405–10,469 | L |
| Asparagine | Asn | N | MT-TN | 5,657–5,729 | H |
| Aspartic acid | Asp | D | MT-TD | 7,518–7,585 | L |
| Cysteine | Cys | C | MT-TC | 5,761–5,826 | H |
| Glutamic acid | Glu | E | MT-TE | 14,674–14,742 | H |
| Glutamine | Gln | Q | MT-TQ | 4,329–4,400 | H |
| Glycine | Gly | G | MT-TG | 9,991–10,058 | L |
| Histidine | His | H | MT-TH | 12,138–12,206 | L |
| Isoleucine | Ile | I | MT-TI | 4,263–4,331 | L |
| Leucine | Leu (UUR) | L | MT-TL1 | 3,230–3,304 | L |
| Leucine | Leu (CUN) | L | MT-TL2 | 12,266–12,336 | L |
| Lysine | Lys | K | MT-TK | 8,295–8,364 | L |
| Methionine | Met | M | MT-TM | 4,402–4,469 | L |
| Phenylalanine | Phe | F | MT-TF | 577–647 | L |
| Proline | Pro | P | MT-TP | 15,956–16,023 | H |
| Serine | Ser (UCN) | S | MT-TS1 | 7,446–7,514 | H |
| Serine | Ser (AGY) | S | MT-TS2 | 12,207–12,265 | L |
| Threonine | Thr | T | MT-TT | 15,888–15,953 | L |
| Tryptophan | Trp | W | MT-TW | 5,512–5,579 | L |
| Tyrosine | Tyr | Y | MT-TY | 5,826–5,891 | H |
| Valine | Val | V | MT-TV | 1,602–1,670 | L |
Location of genes
Mitochondrial
DNA traditionally had the two strands of DNA designated the heavy and
the light strand, due to their buoyant densities during separation in
cesium chloride gradients, which was found to be related to the relative G+T nucleotide content of the strand.
However, confusion of labeling of this strands is widespread, and
appears to originate with an identification of the majority coding
strand as the heavy in one influential article in 1999. In humans, the light strand of mtDNA carries 28 genes and the heavy strand of mtDNA carries only 9 genes.
Eight of the 9 genes on the heavy strand code for mitochondrial tRNA
molecules. Human mtDNA consists of 16,569 nucleotide pairs. The entire
molecule is regulated by only one regulatory region which contains the
origins of replication of both heavy and light strands. The entire human
mitochondrial DNA molecule has been mapped.
Genetic code variants
The genetic code is, for the most part, universal, with few exceptions: mitochondrial genetics includes some of these. For most organisms the "stop codons" are "UAA", "UAG", and "UGA". In vertebrate mitochondria "AGA" and "AGG" are also stop codons, but not "UGA", which codes for tryptophan instead. "AUA" codes for isoleucine in most organisms but for methionine in vertebrate mitochondrial mRNA.
There are many other variations among the codes used by other
mitochondrial m/tRNA, which happened not to be harmful to their
organisms, and which can be used as a tool (along with other mutations
among the mtDNA/RNA of different species) to determine relative
proximity of common ancestry of related species. (The more related two
species are, the more mtDNA/RNA mutations will be the same in their
mitochondrial genome).
Using these techniques, it is estimated that the first mitochondria arose around 1.5 billion years ago. A generally accepted hypothesis is that mitochondria originated as an aerobic prokaryote in a symbiotic relationship within an anaerobic eukaryote.
Replication, repair, transcription, and translation
Mitochondrial replication
is controlled by nuclear genes and is specifically suited to make as
many mitochondria as that particular cell needs at the time.
Mitochondrial transcription in humans is initiated from three promoters,
H1, H2, and L (heavy strand 1, heavy strand 2, and light strand
promoters). The H2 promoter transcribes almost the entire heavy strand
and the L promoter transcribes the entire light strand. The H1 promoter
causes the transcription of the two mitochondrial rRNA molecules.
When transcription takes place on the heavy strand a
polycistronic transcript is created. The light strand produces either
small transcripts, which can be used as primers,
or one long transcript. The production of primers occurs by processing
of light strand transcripts with the Mitochondrial RNase MRP
(Mitochondrial RNA Processing). The requirement of transcription to
produce primers links the process of transcription to mtDNA replication.
Full length transcripts are cut into functional tRNA, rRNA, and mRNA
molecules.
The process of transcription initiation in mitochondria involves three types of proteins: the mitochondrial RNA polymerase (POLRMT), mitochondrial transcription factor A (TFAM), and mitochondrial transcription factors B1 and B2 (TFB1M, TFB2M). POLRMT, TFAM, and TFB1M or TFB2M
assemble at the mitochondrial promoters and begin transcription. The
actual molecular events that are involved in initiation are unknown, but
these factors make up the basal transcription machinery and have been
shown to function in vitro. Mitochondrial translation is still not very well understood. In vitro
translations have still not been successful, probably due to the
difficulty of isolating sufficient mt mRNA, functional mt rRNA, and
possibly because of the complicated changes that the mRNA undergoes
before it is translated.
Mitochondrial DNA polymerase
The Mitochondrial DNA Polymerase (Pol gamma, encoded by the POLG gene) is used in the copying of mtDNA during replication. Because the two (heavy and light) strands on the circular mtDNA molecule have different origins of replication, it replicates in a D-loop mode.
One strand begins to replicate first, displacing the other strand. This
continues until replication reaches the origin of replication on the
other strand, at which point the other strand begins replicating in the
opposite direction. This results in two new mtDNA molecules. Each
mitochondrion has several copies of the mtDNA molecule and the number of
mtDNA molecules is a limiting factor in mitochondrial fission.
After the mitochondrion has enough mtDNA, membrane area, and membrane
proteins, it can undergo fission (very similar to that which bacteria
use) to become two mitochondria. Evidence suggests that mitochondria can
also undergo fusion and exchange (in a form of crossover) genetic material among each other. Mitochondria sometimes form large matrices in which fusion, fission,
and protein exchanges are constantly occurring. mtDNA shared among
mitochondria (despite the fact that they can undergo fusion).
Damage and transcription error
Mitochondrial DNA is susceptible to damage from free oxygen radicals
from mistakes that occur during the production of ATP through the
electron transport chain. These mistakes can be caused by genetic
disorders, cancer, and temperature variations. These radicals can damage
mtDNA molecules or change them, making it hard for mitochondrial
polymerase to replicate them. Both cases can lead to deletions,
rearrangements, and other mutations. Recent evidence has suggested that
mitochondria have enzymes that proofread mtDNA and fix mutations that
may occur due to free radicals. It is believed that a DNA recombinase
found in mammalian cells is also involved in a repairing recombination
process. Deletions and mutations due to free radicals have been
associated with the aging process. It is believed that radicals cause
mutations which lead to mutant proteins, which in turn led to more
radicals. This process takes many years and is associated with some
aging processes involved in oxygen-dependent tissues such as brain,
heart, muscle, and kidney. Auto-enhancing processes such as these are
possible causes of degenerative diseases including Parkinson's, Alzheimer's, and coronary artery disease.
Chromosomally mediated mtDNA replication errors
Because
mitochondrial growth and fission are mediated by the nuclear DNA,
mutations in nuclear DNA can have a wide array of effects on mtDNA
replication. Despite the fact that the loci for some of these mutations
have been found on human chromosomes, specific genes and proteins
involved have not yet been isolated. Mitochondria need a certain protein
to undergo fission. If this protein (generated by the nucleus) is not
present, the mitochondria grow but they do not divide. This leads to
giant, inefficient mitochondria. Mistakes in chromosomal genes or their
products can also affect mitochondrial replication more directly by
inhibiting mitochondrial polymerase and can even cause mutations in the
mtDNA directly and indirectly. Indirect mutations are most often caused
by radicals created by defective proteins made from nuclear DNA.
Mitochondrial diseases
Contribution of mitochondrial versus nuclear genome
In
total, the mitochondrion hosts about 3000 different types of proteins,
but only about 13 of them are coded on the mitochondrial DNA. Most of
the 3000 types of proteins are involved in a variety of processes other
than ATP production, such as porphyrin
synthesis. Only about 3% of them code for ATP production proteins. This
means most of the genetic information coding for the protein makeup of
mitochondria is in chromosomal DNA and is involved in processes other
than ATP synthesis. This increases the chances that a mutation that will
affect a mitochondrion will occur in chromosomal DNA, which is
inherited in a Mendelian pattern. Another result is that a chromosomal
mutation will affect a specific tissue due to its specific needs,
whether those may be high energy requirements or a need for the
catabolism or anabolism of a specific neurotransmitter or nucleic acid.
Because several copies of the mitochondrial genome are carried by each
mitochondrion (2–10 in humans), mitochondrial mutations can be inherited
maternally by mtDNA mutations which are present in mitochondria inside
the oocyte before fertilization, or (as stated above) through mutations in the chromosomes.
Presentation
Mitochondrial diseases
range in severity from asymptomatic to fatal, and are most commonly due
to inherited rather than acquired mutations of mitochondrial DNA. A
given mitochondrial mutation can cause various diseases depending on the
severity of the problem in the mitochondria and the tissue the affected
mitochondria are in. Conversely, several different mutations may
present themselves as the same disease. This almost patient-specific
characterization of mitochondrial diseases (see Personalized medicine)
makes them very hard to accurately recognize, diagnose and trace. Some
diseases are observable at or even before birth (many causing death)
while others do not show themselves until late adulthood (late-onset
disorders). This is because the number of mutant versus wildtype
mitochondria varies between cells and tissues, and is continuously
changing. Because cells have multiple mitochondria, different
mitochondria in the same cell can have different variations of the mtDNA. This condition is referred to as heteroplasmy.
When a certain tissue reaches a certain ratio of mutant versus wildtype
mitochondria, a disease will present itself. The ratio varies from
person to person and tissue to tissue (depending on its specific energy,
oxygen, and metabolism requirements, and the effects of the specific
mutation). Mitochondrial diseases are very numerous and different. Apart
from diseases caused by abnormalities in mitochondrial DNA, many
diseases are suspected to be associated in part by mitochondrial
dysfunctions, such as diabetes mellitus, forms of cancer and cardiovascular disease, lactic acidosis, specific forms of myopathy, osteoporosis, Alzheimer's disease, Parkinsons's disease, stroke, male infertility and which are also believed to play a role in the aging process.
Use in forensics
Human mtDNA can also be used to help identify individuals. Forensic
laboratories occasionally use mtDNA comparison to identify human
remains, and especially to identify older unidentified skeletal remains.
Although unlike nuclear DNA, mtDNA is not specific to one individual,
it can be used in combination with other evidence (anthropological
evidence, circumstantial evidence, and the like) to establish identification. mtDNA is also used to exclude possible matches between missing persons and unidentified remains.
Many researchers believe that mtDNA is better suited to identification
of older skeletal remains than nuclear DNA because the greater number of
copies of mtDNA per cell increases the chance of obtaining a useful
sample, and because a match with a living relative is possible even if
numerous maternal generations separate the two.
Examples
American outlaw Jesse James's
remains were identified using a comparison between mtDNA extracted from
his remains and the mtDNA of the son of the female-line
great-granddaughter of his sister.
Similarly, the remains of Alexandra Feodorovna (Alix of Hesse), last Empress of Russia, and her children were identified by comparison of their mitochondrial DNA with that of Prince Philip, Duke of Edinburgh, whose maternal grandmother was Alexandra's sister Victoria of Hesse.
Similarly to identify Emperor Nicholas II remains his mitochondrial DNA was compared with that of James Carnegie, 3rd Duke of Fife, whose maternal great-grandmother Alexandra of Denmark (Queen Alexandra) was sister of Nicholas II mother Dagmar of Denmark (Empress Maria Feodorovna).
Similarly the remains of king Richard III.