From Wikipedia, the free encyclopedia
Internal symbiont: Mitochondrion has a matrix and membranes, like a free-living proteobacterial cell, from which it may derive.
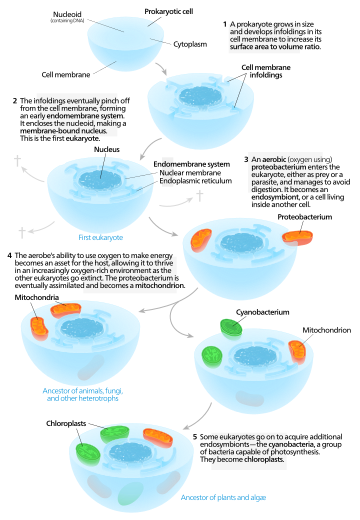
Symbiogenesis, or endosymbiotic theory, is an evolutionary theory of the origin of eukaryotic cells from prokaryotic organisms, first articulated in 1905 and 1910 by the Russian botanist Konstantin Mereschkowski, and advanced and substantiated with microbiological evidence by Lynn Margulis in 1967. It holds that the organelles distinguishing eukaryote cells evolved through symbiosis of individual single-celled prokaryotes (bacteria and archaea).
The theory holds that mitochondria, plastids such as chloroplasts, and possibly other organelles of eukaryotic cells represent formerly free-living prokaryotes taken one inside the other in endosymbiosis, around 1.5 billion years ago. In more detail, mitochondria appear to be related to Rickettsiales proteobacteria, and chloroplasts to nitrogen-fixing filamentous cyanobacteria.
Among the many lines of evidence supporting symbiogenesis are that new mitochondria and plastids are formed only through binary fission, and that cells cannot create new ones otherwise; that the transport proteins called porins are found in the outer membranes of mitochondria, chloroplasts and bacterial cell membranes; that cardiolipin is found only in the inner mitochondrial membrane and bacterial cell membranes; and that some mitochondria and plastids contain single circular DNA molecules similar to the DNA of bacteria.
History
Endosymbiotic theory
Kwang Jeon's experiment supports endosymbiosis. [I] Amoebae infected by
x-bacteria [II] Many amoebae become sick and die [III] Survivors have
x-bacteria living in their cytoplasm [IV] Antibiotics kill x-bacteria:
host amoebae die as now dependent on x-bacteria.
The theory of symbiogenesis (Greek: σύν syn "together", βίωσις biosis "living", and γένεσις genesis "origin or birth") was first articulated by the Russian botanist Konstantin Mereschkowski in his work in 1910, The Theory of Two Plasms as the Basis of Symbiogenesis, a New Study or the Origins of Organisms, although the fundamentals of the idea already had appeared in his earlier 1905 work, The nature and origins of chromatophores in the plant kingdom.[1][2][3] Mereschkowski was familiar with work by botanist Andreas Schimper, who had observed in 1883 that the division of chloroplasts in green plants closely resembled that of free-living cyanobacteria, and who had himself tentatively proposed (in a footnote) that green plants had arisen from a symbiotic union of two organisms.[4] In 1918 the French scientist Paul Portier published Les Symbiotes in which he claimed that the mitochondria originated from a symbiosis process.[5] Ivan Wallin extended the idea of an endosymbiotic origin to mitochondria in the 1920s.[6][7] The Russian botanist Boris Kozo-Polyansky was the first to explain the theory in terms of Darwinian evolution.[8] In his 1924 book Symbiogenesis: A New Principle of Evolution he wrote, "The theory of symbiogenesis is a theory of selection relying on the phenomenon of symbiosis."[9] These theories were initially dismissed or ignored. More detailed electron microscopic comparisons between cyanobacteria and chloroplasts (for example studies by Hans Ris published in 1961[10]), combined with the discovery that plastids and mitochondria contain their own DNA[11] (which by that stage was recognized to be the hereditary material of organisms) led to a resurrection of the idea in the 1960s.
The theory was advanced and substantiated with microbiological evidence by Lynn Margulis in a 1967 paper, On the origin of mitosing cells.[12] In her 1981 work Symbiosis in Cell Evolution she argued that eukaryotic cells originated as communities of interacting entities, including endosymbiotic spirochaetes that developed into eukaryotic flagella and cilia. This last idea has not received much acceptance, because flagella lack DNA and do not show ultrastructural similarities to bacteria or archaea (see also: Evolution of flagella and Prokaryotic cytoskeleton). According to Margulis and Dorion Sagan,[13] "Life did not take over the globe by combat, but by networking" (i.e., by cooperation). The possibility that peroxisomes may have an endosymbiotic origin has also been considered, although they lack DNA. Christian de Duve proposed that they may have been the first endosymbionts, allowing cells to withstand growing amounts of free molecular oxygen in the Earth's atmosphere. However, it now appears that they may be formed de novo, contradicting the idea that they have a symbiotic origin.[14]
It is thought that over millennia these endosymbionts transferred some of their own DNA to the host cell's nucleus (called "endosymbiotic gene transfer" or "endogenosymbiosis") during the evolutionary transition from a symbiotic community to an instituted eukaryotic cell. The endosymbiotic theory is considered to be a type of saltational evolution.[15]
From endosymbionts to organelles
According to Keeling and Archibald,[16] the usual way to distinguish organelles from endosymbionts is by their reduced genome sizes. As an endosymbiont evolves into an organelle, most of their genes are transferred to the host cell genome. The host cell and organelle need to develop a transport mechanism that enables transfer back of the protein products needed by the organelle but now manufactured by the cell. Cyanobacteria and α-proteobacteria are the most closely related free-living organisms to plastids and mitochondria respectively.[17] Both cyanobacteria and α-proteobacteria maintain a large (>6Mb) genome encoding thousands of proteins.[17] Plastids and mitochondria exhibit a dramatic reduction in genome size when compared to their bacterial relatives.[17] Chloroplast genomes in photosynthetic organisms are normally 120-200kb[18] encoding 20-200 proteins[17] and mitochondrial genomes in humans are approximately 16kb and encode 37 genes, 13 of which are proteins.[19] Using the example of the freshwater amoeboid, however, Paulinella chromatophora, which contains chromatophores found to be evolved from cyanobacteria, Keeling and Archibald argue that this is not the only possible criterion; another is that the host cell has assumed control of the regulation of the former endosymbiont's division, thereby synchronizing it with the cell's own division.[16] Nowack and her colleagues[20] performed gene sequencing on the chromatophore (1.02 Mb) and found that only 867 proteins were encoded by these photosynthetic cells. Comparisons with their closest free living cyanobacteria of the genus Synechococcus (having a genome size 3 Mb, with 3300 genes) revealed that chromatophores underwent a drastic genome shrinkage. Chromatophores contained genes that were accountable for photosynthesis but were deficient in genes that could carry out other biosynthetic functions; this observation suggests that these endosymbiotic cells are highly dependent on their hosts for their survival and growth mechanisms. Thus, these chromatophores were found to be non-functional for organelle-specific purposes when compared to mitochondria and plastids. This distinction could have promoted the early evolution of photosynthetic organelles.The loss of genetic autonomy, that is, the loss of many genes from endosymbionts, occurred very early in evolutionary time.[21] Taking into account the entire original endosymbiont genome, there are three main possible fates for genes over evolutionary time. The first fate involves the loss of functionally redundant genes,[21] in which genes that are already represented in the nucleus are eventually lost.The second fate involves the transfer of genes to the nucleus.[17][21][22][23][24] The loss of autonomy and integration of the endosymbiont with its host can be primarily attributed to nuclear gene transfer.[24] As organelle genomes have been greatly reduced over evolutionary time, nuclear genes have expanded and become more complex.[17] As a result, many plastid and mitochondrial processes are driven by nuclear encoded gene products.[17] In addition, many nuclear genes originating from endosymbionts have acquired novel functions unrelated to their organelles.[17][24]
The mechanisms of gene transfer are not fully known; however, multiple hypotheses exist to explain this phenomenon. The cDNA hypothesis involves the use of messenger RNA (mRNAs) to transport genes from organelles to the nucleus where they are converted to cDNA and incorporated into the genome.[17][22] The cDNA hypothesis is based on studies of the genomes of flowering plants.[17] Protein coding RNAs in mitochondria are spliced and edited using organelle-specific splice and editing sites.[17] Nuclear copies of some mitochondrial genes, however, do not contain organelle-specific splice sites, suggesting a processed mRNA intermediate.[17] The cDNA hypothesis has since been revised as edited mitochondrial cDNAs are unlikely to recombine with the nuclear genome and are more likely to recombine with their native mitochondrial genome. If the edited mitochondrial sequence recombines with the mitochondrial genome, mitochondrial splice sites would no longer exist in the mitochondrial genome.[17] Any subsequent nuclear gene transfer would therefore also lack mitochondrial splice sites.[17]
The bulk flow hypothesis is the alternative to the cDNA hypothesis, stating that escaped DNA, rather than mRNA, is the mechanism of gene transfer.[17][22] According to this hypothesis, disturbances to organelles, including autophagy (normal cell destruction), gametogenesis (the formation of gametes), and cell stress, release DNA which is imported into the nucleus and incorporated into the nuclear DNA using non-homologous end joining (repair of double stranded breaks).[22] For example, in the initial stages of endosymbiosis, due to a lack of major gene transfer, the host cell had little to no control over the endosymbiont.[21] The endosymbiont underwent cell division independently of the host cell, resulting in many "copies" of the endosymbiont within the host cell.[21] Some of the endosymbionts lysed (burst), and high levels of DNA were incorporated into the nucleus.[21] A similar mechanism is thought to occur in tobacco plants, who show a high rate of gene transfer and whose cells contain multiple chloroplasts.[21] In addition, the bulk flow hypothesis is also supported by the presence of non-random clusters of organelle genes, suggesting the simultaneous movement of multiple genes.[22]
In 2015, the biologist Roberto Cazzolla Gatti provided evidence for a variant theory,[25] endogenosymbiosis, in which not only are organelles endosymbiotic, but that pieces of genetic material from symbiotic parasites ("gene carriers" such as viruses, retroviruses and bacteriophages), are included in the host's nuclear DNA, changing the host's gene expression and contributing to the process of speciation.[26]
Molecular and biochemical evidence suggests that mitochondria are related to Rickettsiales proteobacteria (in particular, the SAR11 clade,[27][28] or close relatives), and that chloroplasts are related to nitrogen-fixing filamentous cyanobacteria.[29][30]
Organellar genomes
Plastomes and mitogenomes
The human mitochondrial genome has retained genes encoding 2 rRNAs, 22 tRNAs, and 13 redox proteins.
The third and final possible fate of endosymbiont genes is that they remain in the organelles. Plastids and mitochondria, although they have lost much of their genomes, retain genes encoding rRNAs, tRNAs, proteins involved in redox reactions, and proteins required for transcription, translation, and replication.[17][18][21] There are many hypotheses to explain why organelles retain a small portion of their genome; however no one hypothesis will apply to all organisms[21] and the topic is still quite controversial.[17] The hydrophobicity hypothesis states that highly hydrophobic (water hating) proteins (such as the membrane bound proteins involved in redox reactions) are not easily transported through the cytosol and therefore these proteins must be encoded in their respective organelles.[17][21] The code disparity hypothesis states that the limit on transfer is due to differing genetic codes and RNA editing between the organelle and the nucleus.[21] The redox control hypothesis states that genes encoding redox reaction proteins are retained in order to effectively couple the need for repair and the synthesis of these proteins.[17][18][21] For example, if one of the photosystems is lost from the plastid, the intermediate electron carriers may lose or gain too many electrons, signalling the need for repair of a photosystem.[18] The time delay involved in signalling the nucleus and transporting a cytosolic protein to the organelle results in the production of damaging reactive oxygen species.[17][18][21] The final hypothesis states that the assembly of membrane proteins, particularly those involved in redox reactions, requires coordinated synthesis and assembly of subunits; however, translation and protein transport coordination is more difficult to control in the cytoplasm.[21]
Non-photosynthetic plastid genomes
The majority of the genes in the mitochondria and plastids are related to the expression (transcription, translation and replication) of genes encoding proteins involved in either photosynthesis (in plastids) or cellular respiration (in mitochondria).[17][18][21] One might predict, that the loss of photosynthesis or cellular respiration would allow for the complete loss of the plastid genome or the mitochondrial genome respectively.[21] While there are numerous examples of mitochondrial descendants (mitosomes and hydrogenosomes) that have lost their entire organellar genome,[31] non-photosynthetic plastids tend to retain a small genome.[21] There are two main hypotheses to explain this occurrence:The essential tRNA hypothesis notes that there have been no documented functional plastid-to-nucleus gene transfers of genes encoding RNA products (tRNAs and rRNAs).[21] As a result, plastids must make their own functional RNAs or import nuclear counterparts.[21] The genes encoding tRNA-Glu and tRNA-fmet, however, appear to be indispensable.[21] The plastid is responsible for haem biosynthesis, which requires plastid encoded tRNA-Glu (from the gene trnE) as a precursor molecule.[21] Like other genes encoding RNAs, trnE cannot be transferred to the nucleus.[21] In addition, it is unlikely trnE could be replaced by a cytosolic tRNA-Glu as trnE is highly conserved; single base changes in trnE have resulted in the loss of haem synthesis.[21] The gene for tRNA-formylmethionine (tRNA-fmet) is also encoded in the plastid genome and is required for translation initiation in both plastids and mitochondria.[21] A plastid is required to continue expressing the gene for tRNA-fmet so long as the mitochondrion is translating proteins.[21]
The limited window hypothesis offers a more general explanation for the retention of genes in non-photosynthetic plastids.[32] According to the bulk flow hypothesis, genes are transferred to the nucleus following the disturbance of organelles.[22] Disturbance was common in the early stages of endosymbiosis, however, once the host cell gained control of organelle division, eukaryotes could evolve to have only one plastid per cell.[21] Having only one plastid severely limits gene transfer[21] as the lysis of the single plastid would likely result in cell death.[21][32] Consistent with this hypothesis,[32] organisms with multiple plastids show an 80-fold increase in plastid-to-nucleus gene transfer compared to organisms with single plastids.[32]
Evidence
There are many lines of evidence that mitochondria and plastids including chloroplasts arose from bacteria.[33][34][35][36][37]- New mitochondria and plastids are formed only through binary fission, the form of cell division used by bacteria and archaea.[38]
- If a cell's mitochondria or chloroplasts are removed, the cell does not have the means to create new ones.[39] For example, in some algae, such as Euglena, the plastids can be destroyed by certain chemicals or prolonged absence of light without otherwise affecting the cell. In such a case, the plastids will not regenerate.
- Transport proteins called porins are found in the outer membranes of mitochondria and chloroplasts and are also found in bacterial cell membranes.[40][41][42]
- A membrane lipid cardiolipin is exclusively found in the inner mitochondrial membrane and bacterial cell membranes.[43]
- Some mitochondria and some plastids contain single circular DNA molecules that are similar to the DNA of bacteria both in size and structure.[44]
- Genome comparisons suggest a close relationship between mitochondria and Rickettsial bacteria.[45]
- Genome comparisons suggest a close relationship between plastids and cyanobacteria.[46]
- Many genes in the genomes of mitochondria and chloroplasts have been lost or transferred to the nucleus of the host cell. Consequently, the chromosomes of many eukaryotes contain genes that originated from the genomes of mitochondria and plastids.[44]
- Mitochondrial and plastid ribosomes are more similar to those of bacteria (70S) than those of eukaryotes.[47]
- Proteins created by mitochondria and chloroplasts use N-formylmethionine as the initiating amino acid, as do proteins created by bacteria but not proteins created by eukaryotic nuclear genes or archaea.[48][49]
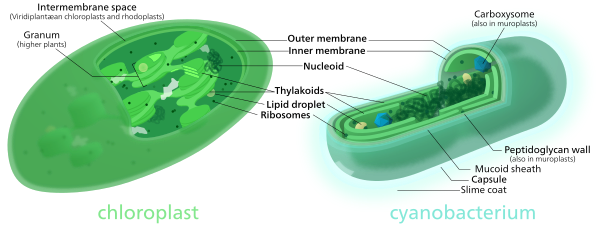
Comparison of chloroplasts and cyanobacteria showing their similarities.
Secondary endosymbiosis
Primary endosymbiosis involves the engulfment of a bacterium by another free living organism. Secondary endosymbiosis occurs when the product of primary endosymbiosis is itself engulfed and retained by another free living eukaryote. Secondary endosymbiosis has occurred several times and has given rise to extremely diverse groups of algae and other eukaryotes. Some organisms can take opportunistic advantage of a similar process, where they engulf an alga and use the products of its photosynthesis, but once the prey item dies (or is lost) the host returns to a free living state. Obligate secondary endosymbionts become dependent on their organelles and are unable to survive in their absence (for a review see McFadden 2001[50]). RedToL, the Red Algal Tree of Life Initiative funded by the National Science Foundation highlights the role red algae or Rhodophyta played in the evolution of our planet through secondary endosymbiosis.One possible secondary endosymbiosis in process has been observed by Okamoto & Inouye (2005). The heterotrophic protist Hatena behaves like a predator until it ingests a green alga, which loses its flagella and cytoskeleton, while Hatena, now a host, switches to photosynthetic nutrition, gains the ability to move towards light and loses its feeding apparatus.[51]
The process of secondary endosymbiosis left its evolutionary signature within the unique topography of plastid membranes. Secondary plastids are surrounded by three (in euglenophytes and some dinoflagellates) or four membranes (in haptophytes, heterokonts, cryptophytes, and chlorarachniophytes). The two additional membranes are thought to correspond to the plasma membrane of the engulfed alga and the phagosomal membrane of the host cell. The endosymbiotic acquisition of a eukaryote cell is represented in the cryptophytes; where the remnant nucleus of the red algal symbiont (the nucleomorph) is present between the two inner and two outer plastid membranes.[citation needed]
Despite the diversity of organisms containing plastids, the morphology, biochemistry, genomic organisation, and molecular phylogeny of plastid RNAs and proteins suggest a single origin of all extant plastids – although this theory is still debated.[52][53]
Some species including Pediculus humanus (lice) have multiple chromosomes in the mitochondrion. This and the phylogenetics of the genes encoded within the mitochondrion suggest that mitochondria have multiple ancestors, that these were acquired by endosymbiosis on several occasions rather than just once, and that there have been extensive mergers and rearrangements of genes on the several original mitochondrial chromosomes.[54]