Studies of heritability ask questions such as how much genetic factors play a role in differences in height between people. This is not the same as asking how much genetic factors influence height in any one person.
Heritability is a statistic used in the fields of breeding and genetics that estimates the degree of variation in a phenotypic trait in a population that is due to genetic variation between individuals in that population.
In other words, the concept of heritability can alternately be
expressed in the form of the following question: "What is the proportion
of the variation in a given trait within a population that is not explained by the environment or random chance?"
Other causes of measured variation in a trait are characterized as environmental factors,
including measurement error. In human studies of heritability these are
often apportioned into factors from "shared environment" and
"non-shared environment" based on whether they tend to result in persons
brought up in the same household being more or less similar to persons
who were not.
Heritability is estimated by comparing individual phenotypic
variation among related individuals in a population. Heritability is an
important concept in quantitative genetics, particularly in selective breeding and behavior genetics (for instance, twin studies).
It is the source of much confusion due to the fact that its technical
definition is different from its commonly-understood folk definition.
Therefore, its use conveys the incorrect impression that behavioral
traits are "inherited" or specifically passed down through the genes.
Behavioral geneticists also conduct heritability analyses based on the
assumption that genes and environments contribute in a separate,
additive manner to behavioral traits.
Overview
Heritability measures the fraction of phenotype variability that can be attributed to genetic variation.
This is not the same as saying that this fraction of an individual
phenotype is caused by genetics. For example, it is incorrect to say
that since the heritability of personality traits is about .6, that
means that 60% of your personality is inherited from your parents and
40% comes from the environment. In addition, heritability can change
without any genetic change occurring, such as when the environment
starts contributing to more variation. As a case in point, consider that
both genes
and environment have the potential to influence intelligence.
Heritability could increase if genetic variation increases, causing
individuals to show more phenotypic variation, like showing different
levels of intelligence. On the other hand, heritability might also
increase if the environmental variation decreases, causing individuals
to show less phenotypic variation, like showing more similar levels of
intelligence. Heritability increases when genetics are contributing more
variation or because non-genetic factors are contributing less
variation; what matters is the relative contribution. Heritability is
specific to a particular population in a particular environment. High
heritability of a trait, consequently, does not necessarily mean that
the trait is not very susceptible to environmental influences. Heritability can also change as a result of changes in the environment, migration, inbreeding, or the way in which heritability itself is measured in the population under study.
The heritability of a trait should not be interpreted as a measure of
the extent to which said trait is genetically determined in an
individual.
The extent of dependence of phenotype on environment can also be a
function of the genes involved. Matters of heritability are complicated
because genes may canalize
a phenotype, making its expression almost inevitable in all occurring
environments. Individuals with the same genotype can also exhibit
different phenotypes through a mechanism called phenotypic plasticity, which makes heritability difficult to measure in some cases. Recent insights in molecular biology have identified changes in transcriptional
activity of individual genes associated with environmental changes.
However, there are a large number of genes whose transcription is not
affected by the environment.
Estimates of heritability use statistical analyses
to help to identify the causes of differences between individuals.
Since heritability is concerned with variance, it is necessarily an
account of the differences between individuals in a population.
Heritability can be univariate
– examining a single trait – or multivariate – examining the genetic
and environmental associations between multiple traits at once. This
allows a test of the genetic overlap between different phenotypes: for
instance hair color and eye color.
Environment and genetics may also interact, and heritability analyses
can test for and examine these interactions (GxE models).
A prerequisite for heritability analyses is that there is some
population variation to account for. This last point highlights the fact
that heritability cannot take into account the effect of factors which
are invariant in the population. Factors may be invariant if they are
absent and do not exist in the population, such as no one having access
to a particular antibiotic, or because they are omni-present, like if everyone is drinking coffee. In practice, all human behavioral traits vary and almost all traits show some heritability.
Definition
Any particular phenotype can be modeled as the sum of genetic and environmental effects:
- Phenotype (P) = Genotype (G) + Environment (E).
Likewise the phenotypic variance in the trait – Var (P) – is the sum of effects as follows:
- Var(P) = Var(G) + Var(E) + 2 Cov(G,E).
In a planned experiment Cov(G,E) can be controlled and held at 0. In this case, heritability is defined as:
- .
H2 is the broad-sense heritability. This reflects
all the genetic contributions to a population's phenotypic variance
including additive, dominant, and epistatic (multi-genic interactions), as well as maternal and paternal effects, where individuals are directly affected by their parents' phenotype, such as with milk production in mammals.
A particularly important component of the genetic variance is the
additive variance, Var(A), which is
the variance due to the average
effects (additive effects) of the alleles. Since each parent passes a single allele per locus
to each offspring, parent-offspring resemblance depends upon the
average effect of single alleles. Additive variance represents,
therefore, the genetic component of variance responsible for
parent-offspring resemblance. The additive genetic portion of the
phenotypic variance is known as Narrow-sense heritability and is defined
as
An upper case H2 is used to denote broad sense, and lower case h2 for narrow sense.
For traits which are not continuous but dichotomous such as an
additional toe or certain diseases, the contribution of the various
alleles can be considered to be a sum, which past a threshold, manifests
itself as the trait, giving the liability threshold model in which heritability can be estimated and selection modeled.
Additive variance is important for selection.
If a selective pressure such as improving livestock is exerted, the
response of the trait is directly related to narrow-sense heritability.
The mean of the trait will increase in the next generation as a function
of how much the mean of the selected parents differs from the mean of
the population from which the selected parents were chosen. The observed
response to selection leads to an estimate of the narrow-sense heritability (called realized heritability). This is the principle underlying artificial selection or breeding.
Example
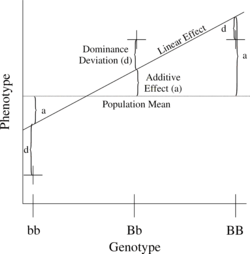
Figure 1. Relationship of phenotypic values to additive and dominance effects using a completely dominant locus.
The simplest genetic model involves a single locus with two alleles (b and B) affecting one quantitative phenotype.
The number of B alleles can vary from 0, 1, or 2. For any genotype, BiBj, the expected phenotype can then be written as the sum of the overall mean, a linear effect, and a dominance deviation:
- = Population mean +
- Additive Effect () +
- Dominance Deviation ().
The additive genetic variance at this locus is the weighted average of the squares of the additive effects:
where
There is a similar relationship for variance of dominance deviations:
where
The linear regression of phenotype on genotype is shown in Figure 1.
Assumptions
Estimates
of the total heritability of human traits assume the absence of
epistasis, which has been called the "assumption of additivity".
Although some researchers have cited such estimates in support of the
existence of "missing heritability" unaccounted for by known genetic loci, the assumption of additivity may render these estimates invalid.
There is also some empirical evidence that the additivity assumption is
frequently violated in behavior genetic studies of adolescent
intelligence and academic achievement.
Estimating heritability
Since only P
can be observed or measured directly, heritability must be estimated
from the similarities observed in subjects varying in their level of
genetic or environmental similarity. The statistical analyses required to estimate the genetic and environmental
components of variance depend on the sample characteristics. Briefly,
better estimates are obtained using data from individuals with widely
varying levels of genetic relationship - such as twins, siblings, parents and offspring, rather than from more distantly related (and therefore less similar) subjects. The standard error for heritability estimates is improved with large sample sizes.
In non-human populations it is often possible to collect
information in a controlled way. For example, among farm animals it is
easy to arrange for a bull to produce offspring from a large number of
cows and to control environments. Such experimental control is generally not possible when gathering human data, relying on naturally occurring relationships and environments.
In classical quantitative genetics, there were two schools of thought regarding estimation of heritability.
One school of thought was developed by Sewall Wright at The University of Chicago, and further popularized by C. C. Li (University of Chicago) and J. L. Lush (Iowa State University). It is based on the analysis of correlations and, by extension, regression. Path Analysis was developed by Sewall Wright as a way of estimating heritability.
The second was originally developed by R. A. Fisher and expanded at The University of Edinburgh, Iowa State University, and North Carolina State University, as well as other schools. It is based on the analysis of variance
of breeding studies, using the intraclass correlation of relatives.
Various methods of estimating components of variance (and, hence,
heritability) from ANOVA are used in these analyses.
Today, heritability can be estimated from general pedigrees using linear mixed models and from genomic relatedness estimated from genetic markers.
Studies of human heritability often utilize adoption study designs, often with identical twins
who have been separated early in life and raised in different
environments. Such individuals have identical genotypes and can be used
to separate the effects of genotype and environment. A limit of this
design is the common prenatal environment and the relatively low numbers
of twins reared apart. A second and more common design is the twin study
in which the similarity of identical and fraternal twins is used to
estimate heritability. These studies can be limited by the fact that
identical twins are not completely genetically identical, potentially resulting in an underestimation of heritability.
In observational studies, or because of evocative effects (where a genome evokes environments by its effect on them), G and E may covary: gene environment correlation.
Depending on the methods used to estimate heritability, correlations
between genetic factors and shared or non-shared environments may or may
not be confounded with heritability.
Regression/correlation methods of estimation
The first school of estimation uses regression and correlation to estimate heritability.
Comparison of close relatives
In the comparison of relatives, we find that in general,
where r can be thought of as the coefficient of relatedness, b is the coefficient of regression and t is the coefficient of correlation.
Parent-offspring regression
Figure 2. Sir Francis Galton's
(1889) data showing the relationship between offspring height (928
individuals) as a function of mean parent height (205 sets of parents).
Heritability may be estimated by comparing parent and offspring
traits (as in Fig. 2). The slope of the line (0.57) approximates the
heritability of the trait when offspring values are regressed against
the average trait in the parents. If only one parent's value is used
then heritability is twice the slope. (Note that this is the source of
the term "regression," since the offspring values always tend to regress to the mean value for the population, i.e., the slope is always less than one). This regression effect also underlies the DeFries–Fulker method for analyzing twins selected for one member being affected.
Sibling comparison
A
basic approach to heritability can be taken using full-Sib designs:
comparing similarity between siblings who share both a biological mother
and a father. When there is only additive gene action, this sibling phenotypic correlation is an index of familiarity
– the sum of half the additive genetic variance plus full effect of the
common environment. It thus places an upper-limit on additive
heritability of twice the full-Sib phenotypic correlation. Half-Sib
designs compare phenotypic traits of siblings that share one parent with
other sibling groups.
Twin studies
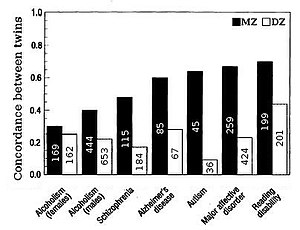
Figure
3. Twin concordances for seven psychological traits (sample size shown
inside bars), with DZ being fraternal and MZ being identical twins.
Heritability for traits in humans is most frequently estimated by
comparing resemblances between twins. "The advantage of twin studies, is
that the total variance can be split up into genetic, shared or common
environmental, and unique environmental components, enabling an accurate
estimation of heritability". Fraternal or dizygotic (DZ) twins on average share half their genes (assuming there is no assortative mating
for the trait), and so identical or monozygotic (MZ) twins on average
are twice as genetically similar as DZ twins. A crude estimate of
heritability, then, is approximately twice the difference in correlation between MZ and DZ twins, i.e. Falconer's formula H2=2(r(MZ)-r(DZ)).
The effect of shared environment, c2,
contributes to similarity between siblings due to the commonality of the
environment they are raised in. Shared environment is approximated by
the DZ correlation minus half heritability, which is the degree to which
DZ twins share the same genes, c2=DZ-1/2h2. Unique environmental variance, e2, reflects the degree to which identical twins raised together are dissimilar, e2=1-r(MZ).
Analysis of variance methods of estimation
The second set of methods of estimation of heritability involves ANOVA and estimation of variance components.
Basic model
We use the basic discussion of Kempthorne.
Considering only the most basic of genetic models, we can look at the
quantitative contribution of a single locus with genotype Gi as
where is the effect of genotype Gi and is the environmental effect.
Consider an experiment with a group of sires and their progeny
from random dams. Since the progeny get half of their genes from the
father and half from their (random) mother, the progeny equation is
Intraclass correlations
Consider
the experiment above. We have two groups of progeny we can compare. The
first is comparing the various progeny for an individual sire (called within sire group).
The variance will include terms for genetic variance (since they did
not all get the same genotype) and environmental variance. This is
thought of as an error term.
The second group of progeny are comparisons of means of half sibs with each other (called among sire group). In addition to the error term
as in the within sire groups, we have an addition term due to the
differences among different means of half sibs. The intraclass
correlation is
- ,
since environmental effects are independent of each other.
The ANOVA
In an experiment with sires and progeny per sire, we can calculate the following ANOVA, using as the genetic variance and as the environmental variance:
| Source | d.f. | Mean Square | Expected Mean Square |
|---|---|---|---|
| Among sire groups | |||
| Within sire groups |
The term is the intraclass correlation among half sibs. We can easily calculate .
The Expected Mean Square is calculated from the relationship of the
individuals (progeny within a sire are all half-sibs, for example), and
an understanding of intraclass correlations.
The use of ANOVA to calculate heritability often fails to account for the presence of gene-environment interactions, because ANOVA has a much lower statistical power for testing for interaction effects than for direct effects.
Model with additive and dominance terms
For a model with additive and dominance terms, but not others, the equation for a single locus is
where is the additive effect of the ith allele, is the additive effect of the jth allele, is the dominance deviation for the ijth genotype, and is the environment.
Experiments can be run with a similar setup to the one given in
Table 1. Using different relationship groups, we can evaluate different
intraclass correlations. Using as the additive genetic variance and as the dominance deviation variance, intraclass correlations become linear functions of these parameters. In general,
- Intraclass correlation
where and are found as
Some common relationships and their coefficients are given in Table 2.
| Relationship | ||
|---|---|---|
| Identical Twins | ||
| Parent-Offspring | ||
| Half Siblings | ||
| Full Siblings | ||
| First Cousins | ||
| Double First Cousins |
Linear mixed models
A
wide variety approaches using linear mixed models have been reported in
literature. Via these methods, phenotypic variance is partitioned into
genetic, environmental and experimental design variances to estimate
heritability. Environmental variance can be explicitly modeled by
studying individuals across a broad range of environments, although
inference of genetic variance from phenotypic and environmental variance
may lead to underestimation of heritability due to the challenge of
capturing the full range of environmental influence affecting a trait.
Other methods for calculating heritability use data from genome-wide association studies
to estimate the influence on a trait by genetic factors, which is
reflected by the rate and influence of putatively associated genetic
loci (usually single-nucleotide polymorphisms)
on the trait. This can lead to underestimation of heritability,
however. This discrepancy is referred to as "missing heritability" and
reflects the challenge of accurately modeling both genetic and
environmental variance in heritability models.
When a large, complex pedigree or another aforementioned type of
data is available, heritability and other quantitative genetic
parameters can be estimated by restricted maximum likelihood (REML) or Bayesian methods. The raw data
will usually have three or more data points for each individual: a code
for the sire, a code for the dam and one or several trait values.
Different trait values may be for different traits or for different time
points of measurement.
The currently popular methodology relies on high degrees of
certainty over the identities of the sire and dam; it is not common to
treat the sire identity probabilistically. This is not usually a
problem, since the methodology is rarely applied to wild populations
(although it has been used for several wild ungulate and bird
populations), and sires are invariably known with a very high degree of
certainty in breeding programmes. There are also algorithms that account
for uncertain paternity.
The pedigrees can be viewed using programs such as Pedigree Viewer , and analyzed with programs such as ASReml, VCE , WOMBAT or the BLUPF90 family of programs.
Pedigree models are helpful for untangling confounds such as reverse causality, maternal effects such as the prenatal environment, and confounding of genetic dominance, shared environment, and maternal gene effects.
Genomic heritability
When
genome-wide genotype data and phenotypes from large population samples
are available, one can estimate the relationships between individuals
based on their genotypes and use a linear mixed model to estimate the
variance explained by the genetic markers. This gives a genomic
heritability estimate based on the variance captured by common genetic
variants. There are multiple methods that make different adjustments for allele frequency and linkage disequilibrium.
Response to selection
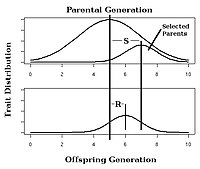
Figure 4. Strength of selection (S) and response to selection (R) in an artificial selection experiment, h2=R/S.
In selective breeding of plants and animals, the expected response to selection of a trait with known narrow-sense heritability can be estimated using the breeder's equation:
In this equation, the Response to Selection (R) is defined as the
realized average difference between the parent generation and the next
generation, and the Selection Differential (S) is defined as the average
difference between the parent generation and the selected parents.
For example, imagine that a plant breeder is involved in a
selective breeding project with the aim of increasing the number of
kernels per ear of corn. For the sake of argument, let us assume that
the average ear of corn in the parent generation has 100 kernels. Let us
also assume that the selected parents produce corn with an average of
120 kernels per ear. If h2 equals 0.5, then the next
generation will produce corn with an average of 0.5(120-100) = 10
additional kernels per ear. Therefore, the total number of kernels per
ear of corn will equal, on average, 110.
Observing the response to selection in an artificial selection
experiment will allow calculation of realized heritability as in Fig. 5.
Note that heritability in the above equation is equal to the ratio only if the genotype and the environmental noise follow Gaussian distributions.
Controversies
Heritability estimates' prominent critics, such as Steven Rose, Jay Joseph, and Richard Bentall, focus largely on heritability estimates in behavioral sciences and social sciences.
Bentall has claimed that such heritability scores are typically
calculated counterintuitively to derive numerically high scores, that
heritability is misinterpreted as genetic determination,
and that this alleged bias distracts from other factors that researches
have found more causally important, such as childhood abuse causing
later psychosis.
Heritability estimates are also inherently limited because they do not
convey any information regarding whether genes or environment play a
larger role in the development of the trait under study. For this
reason, David Moore and David Shenk
describe the term "heritability" in the context of behavior genetics as
"...one of the most misleading in the history of science" and argue
that it has no value except in very rare cases.
When studying complex human traits, it is impossible to use
heritability analysis to determine the relative contributions of genes
and environment, as such traits result from multiple causes interacting.
The controversy over heritability estimates is largely via their basis in twin studies. The scarce success of molecular-genetic studies to corroborate such population-genetic studies' conclusions is the missing heritability problem. Eric Turkheimer has argued that newer molecular methods have vindicated the conventional interpretation of twin studies, although it remains mostly unclear how to explain the relations between genes and behaviors.
According to Turkheimer, both genes and environment are heritable,
genetic contribution varies by environment, and a focus on heritability
distracts from other important factors. Overall, however, heritability is a concept widely applicable.