Diagram of molecular cloning using bacteria and plasmids.
Molecular cloning is a set of experimental methods in molecular biology that are used to assemble recombinant DNA molecules and to direct their replication within host organisms. The use of the word cloning
refers to the fact that the method involves the replication of one
molecule to produce a population of cells with identical DNA molecules.
Molecular cloning generally uses DNA sequences from two different
organisms: the species that is the source of the DNA to be cloned, and
the species that will serve as the living host
for replication of the recombinant DNA. Molecular cloning methods are
central to many contemporary areas of modern biology and medicine.
In a conventional molecular cloning experiment, the DNA to be
cloned is obtained from an organism of interest, then treated with
enzymes in the test tube to generate smaller DNA fragments.
Subsequently, these fragments are then combined with vector DNA
to generate recombinant DNA molecules. The recombinant DNA is then
introduced into a host organism (typically an easy-to-grow, benign,
laboratory strain of E. coli
bacteria). This will generate a population of organisms in which
recombinant DNA molecules are replicated along with the host DNA.
Because they contain foreign DNA fragments, these are transgenic or genetically modified microorganisms (GMO).
This process takes advantage of the fact that a single bacterial cell
can be induced to take up and replicate a single recombinant DNA
molecule. This single cell can then be expanded exponentially to
generate a large amount of bacteria, each of which contain copies of the
original recombinant molecule. Thus, both the resulting bacterial
population, and the recombinant DNA molecule, are commonly referred to
as "clones". Strictly speaking, recombinant DNA refers to DNA molecules, while molecular cloning
refers to the experimental methods used to assemble them. The idea
arose that different DNA sequences could be inserted into a plasmid and
that these foreign sequences would be carried into bacteria and digested
as part of the plasmid. That is, these plasmids could serve as cloning
vectors to carry genes.
Virtually any DNA sequence can be cloned and amplified, but there
are some factors that might limit the success of the process. Examples
of the DNA sequences that are difficult to clone are inverted repeats,
origins of replication, centromeres and telomeres. Another
characteristic that limits chances of success is large size of DNA
sequence. Inserts larger than 10kbp have very limited success, but
bacteriophages such as bacteriophage λ can be modified to successfully
insert a sequence up to 40 kbp.
History
Prior
to the 1970s, our understanding of genetics and molecular biology was
severely hampered by an inability to isolate and study individual genes
from complex organisms. This changed dramatically with the advent of
molecular cloning methods. Microbiologists, seeking to understand the
molecular mechanisms through which bacteria restricted the growth of
bacteriophage, isolated restriction endonucleases, enzymes that could cleave DNA molecules only when specific DNA sequences were encountered.
They showed that restriction enzymes cleaved chromosome-length DNA
molecules at specific locations, and that specific sections of the
larger molecule could be purified by size fractionation. Using a second
enzyme, DNA ligase, fragments generated by restriction enzymes could be joined in new combinations, termed recombinant DNA.
By recombining DNA segments of interest with vector DNA, such as
bacteriophage or plasmids, which naturally replicate inside bacteria,
large quantities of purified recombinant DNA molecules could be produced
in bacterial cultures. The first recombinant DNA molecules were
generated and studied in 1972.
Overview
Molecular cloning takes advantage of the fact that the chemical structure of DNA
is fundamentally the same in all living organisms. Therefore, if any
segment of DNA from any organism is inserted into a DNA segment
containing the molecular sequences required for DNA replication, and the resulting recombinant DNA
is introduced into the organism from which the replication sequences
were obtained, then the foreign DNA will be replicated along with the
host cell's DNA in the transgenic organism.
Molecular cloning is similar to polymerase chain reaction
(PCR) in that it permits the replication of DNA sequence. The
fundamental difference between the two methods is that molecular cloning
involves replication of the DNA in a living microorganism, while PCR
replicates DNA in an in vitro solution, free of living cells.
Steps
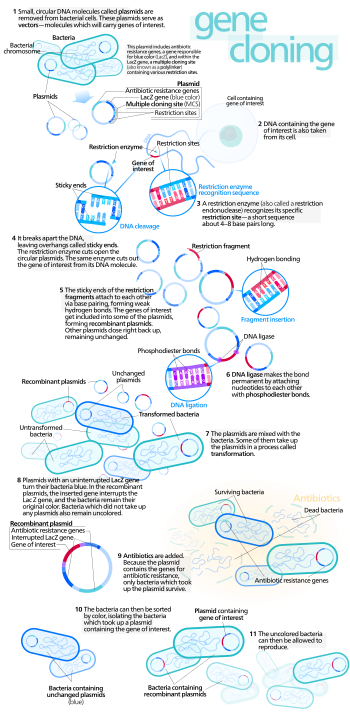
The overall goal of molecular cloning is to take a gene of interest from one plasmid and insert it into another plasmid This is done by performing PCR, digestive reaction, ligation reaction, and transformation.
In standard molecular cloning experiments, the cloning of any DNA
fragment essentially involves seven steps: (1) Choice of host organism
and cloning vector, (2) Preparation of vector DNA, (3) Preparation of
DNA to be cloned, (4) Creation of recombinant DNA, (5) Introduction of
recombinant DNA into host organism, (6) Selection of organisms
containing recombinant DNA, (7) Screening for clones with desired DNA
inserts and biological properties.
Although the detailed planning of the cloning can be done in any
text editor, together with online utilities for e.g. PCR primer design,
dedicated software exist for the purpose. Software for the purpose
include for example ApE (open source), DNAStrider (open source), Serial Cloner (gratis) and Collagene (open source).
Notably, the growing capacity and fidelity of DNA synthesis
platforms allows for increasingly intricate designs in molecular
engineering. These projects may include very long strands of novel DNA
sequence and/or test entire libraries simultaneously, as opposed to of
individual sequences. These shifts introduce complexity that require
design to move away from the flat nucleotide-based representation and
towards a higher level of abstraction. Examples of such tools are GenoCAD, Teselagen (free for academia) or GeneticConstructor (free for academics).
Choice of host organism and cloning vector
Diagram of a commonly used cloning plasmid; pBR322. It's a circular piece of DNA 4361 bases long. Two antibiotic resistance genes are present, conferring resistance to ampicillin and tetracycline, and an origin of replication that the host uses to replicate the DNA.
Although a very large number of host organisms and molecular cloning
vectors are in use, the great majority of molecular cloning experiments
begin with a laboratory strain of the bacterium E. coli (Escherichia coli) and a plasmid cloning vector. E. coli
and plasmid vectors are in common use because they are technically
sophisticated, versatile, widely available, and offer rapid growth of
recombinant organisms with minimal equipment. If the DNA to be cloned is exceptionally large (hundreds of thousands to millions of base pairs), then a bacterial artificial chromosome or yeast artificial chromosome vector is often chosen.
Specialized applications may call for specialized host-vector
systems. For example, if the experimentalists wish to harvest a
particular protein from the recombinant organism, then an expression vector
is chosen that contains appropriate signals for transcription and
translation in the desired host organism. Alternatively, if replication
of the DNA in different species is desired (for example, transfer of DNA
from bacteria to plants), then a multiple host range vector (also
termed shuttle vector)
may be selected. In practice, however, specialized molecular cloning
experiments usually begin with cloning into a bacterial plasmid,
followed by subcloning into a specialized vector.
Whatever combination of host and vector are used, the vector
almost always contains four DNA segments that are critically important
to its function and experimental utility:
- DNA replication origin is necessary for the vector (and its linked recombinant sequences) to replicate inside the host organism
- one or more unique restriction endonuclease recognition sites to serves as sites where foreign DNA may be introduced
- a selectable genetic marker gene that can be used to enable the survival of cells that have taken up vector sequences
- a tag gene that can be used to screen for cells containing the foreign DNA
Cleavage of a DNA sequence containing the BamHI restriction site. The DNA is cleaved at the palindromic sequence to produce 'sticky ends'.
Preparation of vector DNA
The
cloning vector is treated with a restriction endonuclease to cleave the
DNA at the site where foreign DNA will be inserted. The restriction
enzyme is chosen to generate a configuration at the cleavage site that
is compatible with the ends of the foreign DNA (see DNA end). Typically, this is done by cleaving the vector DNA and foreign DNA with the same restriction enzyme, for example EcoRI.
Most modern vectors contain a variety of convenient cleavage sites that
are unique within the vector molecule (so that the vector can only be
cleaved at a single site) and are located within a gene (frequently beta-galactosidase)
whose inactivation can be used to distinguish recombinant from
non-recombinant organisms at a later step in the process. To improve the
ratio of recombinant to non-recombinant organisms, the cleaved vector
may be treated with an enzyme (alkaline phosphatase)
that dephosphorylates the vector ends. Vector molecules with
dephosphorylated ends are unable to replicate, and replication can only
be restored if foreign DNA is integrated into the cleavage site.
Preparation of DNA to be cloned
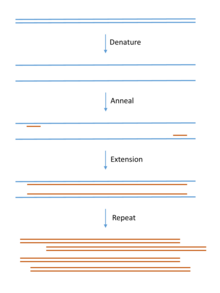
DNA for cloning is most commonly produced using PCR. Template DNA is mixed with bases (the building blocks of DNA), primers (short pieces of complementary single stranded DNA) and a DNA polymerase enzyme that builds the DNA chain. The mix goes through cycles of heating and cooling to produce large quantities of copied DNA.
For cloning of genomic DNA, the DNA to be cloned is extracted from
the organism of interest. Virtually any tissue source can be used (even
tissues from extinct animals),
as long as the DNA is not extensively degraded. The DNA is then
purified using simple methods to remove contaminating proteins
(extraction with phenol), RNA (ribonuclease) and smaller molecules
(precipitation and/or chromatography). Polymerase chain reaction (PCR) methods are often used for amplification of specific DNA or RNA (RT-PCR) sequences prior to molecular cloning.
DNA for cloning experiments may also be obtained from RNA using reverse transcriptase (complementary DNA or cDNA cloning), or in the form of synthetic DNA (artificial gene synthesis).
cDNA cloning is usually used to obtain clones representative of the
mRNA population of the cells of interest, while synthetic DNA is used to
obtain any precise sequence defined by the designer.
The purified DNA is then treated with a restriction enzyme to
generate fragments with ends capable of being linked to those of the
vector. If necessary, short double-stranded segments of DNA (linkers) containing desired restriction sites may be added to create end structures that are compatible with the vector.
Creation of recombinant DNA with DNA ligase
The
creation of recombinant DNA is in many ways the simplest step of the
molecular cloning process. DNA prepared from the vector and foreign
source are simply mixed together at appropriate concentrations and
exposed to an enzyme (DNA ligase) that covalently links the ends together. This joining reaction is often termed ligation. The resulting DNA mixture containing randomly joined ends is then ready for introduction into the host organism.
DNA ligase only recognizes and acts on the ends of linear DNA
molecules, usually resulting in a complex mixture of DNA molecules with
randomly joined ends. The desired products (vector DNA covalently linked
to foreign DNA) will be present, but other sequences (e.g. foreign DNA
linked to itself, vector DNA linked to itself and higher-order
combinations of vector and foreign DNA) are also usually present. This
complex mixture is sorted out in subsequent steps of the cloning
process, after the DNA mixture is introduced into cells.
Introduction of recombinant DNA into host organism
The
DNA mixture, previously manipulated in vitro, is moved back into a
living cell, referred to as the host organism. The methods used to get
DNA into cells are varied, and the name applied to this step in the
molecular cloning process will often depend upon the experimental method
that is chosen (e.g. transformation, transduction, transfection, electroporation).
When microorganisms are able to take up and replicate DNA from their local environment, the process is termed transformation, and cells that are in a physiological state such that they can take up DNA are said to be competent. In mammalian cell culture, the analogous process of introducing DNA into cells is commonly termed transfection.
Both transformation and transfection usually require preparation of the
cells through a special growth regime and chemical treatment process
that will vary with the specific species and cell types that are used.
Electroporation uses high voltage electrical pulses to translocate DNA across the cell membrane (and cell wall, if present). In contrast, transduction
involves the packaging of DNA into virus-derived particles, and using
these virus-like particles to introduce the encapsulated DNA into the
cell through a process resembling viral infection. Although
electroporation and transduction are highly specialized methods, they
may be the most efficient methods to move DNA into cells.
Selection of organisms containing vector sequences
Whichever
method is used, the introduction of recombinant DNA into the chosen
host organism is usually a low efficiency process; that is, only a small
fraction of the cells will actually take up DNA. Experimental
scientists deal with this issue through a step of artificial genetic
selection, in which cells that have not taken up DNA are selectively
killed, and only those cells that can actively replicate DNA containing
the selectable marker gene encoded by the vector are able to survive.
When bacterial cells are used as host organisms, the selectable marker is usually a gene that confers resistance to an antibiotic that would otherwise kill the cells, typically ampicillin.
Cells harboring the plasmid will survive when exposed to the
antibiotic, while those that have failed to take up plasmid sequences
will die. When mammalian cells (e.g. human or mouse cells) are used, a
similar strategy is used, except that the marker gene (in this case
typically encoded as part of the kanMX cassette) confers resistance to the antibiotic Geneticin.
Screening for clones with desired DNA inserts and biological properties
Modern bacterial cloning vectors (e.g. pUC19 and later derivatives including the pGEM vectors) use the blue-white screening system
to distinguish colonies (clones) of transgenic cells from those that
contain the parental vector (i.e. vector DNA with no recombinant
sequence inserted). In these vectors, foreign DNA is inserted into a
sequence that encodes an essential part of beta-galactosidase,
an enzyme whose activity results in formation of a blue-colored colony
on the culture medium that is used for this work. Insertion of the
foreign DNA into the beta-galactosidase coding sequence disables the
function of the enzyme, so that colonies containing transformed DNA
remain colorless (white). Therefore, experimentalists are easily able to
identify and conduct further studies on transgenic bacterial clones,
while ignoring those that do not contain recombinant DNA.
The total population of individual clones obtained in a molecular cloning experiment is often termed a DNA library.
Libraries may be highly complex (as when cloning complete genomic DNA
from an organism) or relatively simple (as when moving a previously
cloned DNA fragment into a different plasmid), but it is almost always
necessary to examine a number of different clones to be sure that the
desired DNA construct is obtained. This may be accomplished through a
very wide range of experimental methods, including the use of nucleic acid hybridizations, antibody probes, polymerase chain reaction, restriction fragment analysis and/or DNA sequencing.
Applications
Molecular
cloning provides scientists with an essentially unlimited quantity of
any individual DNA segments derived from any genome. This material can
be used for a wide range of purposes, including those in both basic and
applied biological science. A few of the more important applications are
summarized here.
Genome organization and gene expression
Molecular
cloning has led directly to the elucidation of the complete DNA
sequence of the genomes of a very large number of species and to an
exploration of genetic diversity within individual species, work that
has been done mostly by determining the DNA sequence of large numbers of
randomly cloned fragments of the genome, and assembling the overlapping
sequences.
At the level of individual genes, molecular clones are used to generate probes that are used for examining how genes are expressed,
and how that expression is related to other processes in biology,
including the metabolic environment, extracellular signals, development,
learning, senescence and cell death. Cloned genes can also provide
tools to examine the biological function and importance of individual
genes, by allowing investigators to inactivate the genes, or make more subtle mutations using regional mutagenesis or site-directed mutagenesis. Genes cloned into expression vectors for functional cloning provide a means to screen for genes on the basis of the expressed protein's function.
Production of recombinant proteins
Obtaining
the molecular clone of a gene can lead to the development of organisms
that produce the protein product of the cloned genes, termed a
recombinant protein. In practice, it is frequently more difficult to
develop an organism that produces an active form of the recombinant
protein in desirable quantities than it is to clone the gene. This is
because the molecular signals for gene expression are complex and
variable, and because protein folding, stability and transport can be
very challenging.
Many useful proteins are currently available as recombinant products.
These include--(1) medically useful proteins whose administration can
correct a defective or poorly expressed gene (e.g. recombinant factor VIII, a blood-clotting factor deficient in some forms of hemophilia, and recombinant insulin, used to treat some forms of diabetes), (2) proteins that can be administered to assist in a life-threatening emergency (e.g. tissue plasminogen activator, used to treat strokes),
(3) recombinant subunit vaccines, in which a purified protein can be
used to immunize patients against infectious diseases, without exposing
them to the infectious agent itself (e.g. hepatitis B vaccine), and (4) recombinant proteins as standard material for diagnostic laboratory tests.
Transgenic organisms
Once
characterized and manipulated to provide signals for appropriate
expression, cloned genes may be inserted into organisms, generating
transgenic organisms, also termed genetically modified organisms (GMOs). Although most GMOs are generated for purposes of basic biological research,
a number of GMOs have been developed for commercial use, ranging from
animals and plants that produce pharmaceuticals or other compounds (pharming), herbicide-resistant crop plants, and fluorescent tropical fish (GloFish) for home entertainment.
Gene therapy
Gene
therapy involves supplying a functional gene to cells lacking that
function, with the aim of correcting a genetic disorder or acquired
disease. Gene therapy can be broadly divided into two categories. The
first is alteration of germ cells, that is, sperm or eggs, which results
in a permanent genetic change for the whole organism and subsequent
generations. This “germ line gene therapy” is considered by many to be
unethical in human beings.
The second type of gene therapy, “somatic cell gene therapy”, is
analogous to an organ transplant. In this case, one or more specific
tissues are targeted by direct treatment or by removal of the tissue,
addition of the therapeutic gene or genes in the laboratory, and return
of the treated cells to the patient. Clinical trials of somatic cell
gene therapy began in the late 1990s, mostly for the treatment of
cancers and blood, liver, and lung disorders.
Despite a great deal of publicity and promises, the history of
human gene therapy has been characterized by relatively limited success.
The effect of introducing a gene into cells often promotes only partial
and/or transient relief from the symptoms of the disease being treated.
Some gene therapy trial patients have suffered adverse consequences of
the treatment itself, including deaths. In some cases, the adverse
effects result from disruption of essential genes within the patient's
genome by insertional inactivation. In others, viral vectors used for
gene therapy have been contaminated with infectious virus. Nevertheless,
gene therapy is still held to be a promising future area of medicine,
and is an area where there is a significant level of research and
development activity.