Biomedical text mining (including biomedical natural language processing or BioNLP) refers to the methods and study of how text mining may be applied to texts and literature of the biomedical and molecular biology domains. As a field of research, biomedical text mining incorporates ideas from natural language processing, bioinformatics, medical informatics and computational linguistics. The strategies developed through studies in this field are frequently applied to the biomedical and molecular biology literature available through services such as PubMed.
Considerations
Applying text mining approaches to biomedical text requires specific considerations common to the domain.
Availability of annotated text data
Large annotated corpora used in the development and training of general purpose text mining methods (e.g., sets of movie dialogue, product reviews, or Wikipedia article text) are not specific for biomedical language. While they may provide evidence of general text properties such as parts of speech, they rarely contain concepts of interest to biologists or clinicians. Development of new methods to identify features specific to biomedical documents therefore requires assembly of specialized corpora. Resources designed to aid in building new biomedical text mining methods have been developed through the Informatics for Integrating Biology and the Bedside (i2b2) challenges and biomedical informatics researchers. Text mining researchers frequently combine these corpora with the controlled vocabularies and ontologies available through the National Library of Medicine's Unified Medical Language System (UMLS) and Medical Subject Headings (MeSH).
Machine learning-based methods often require very large data sets as training data to build useful models. Manual annotation of large text corpora is not realistically possible. Training data may therefore be products of weak supervision or purely statistical methods.
Data structure variation
Like other text documents, biomedical documents contain unstructured data. Research publications follow different formats, contain different types of information, and are interspersed with figures, tables, and other non-text content. Both unstructured text and semi-structured document elements, such as tables, may contain important information that should be text mined. Clinical documents may vary in structure and language between departments and locations. Other types of biomedical text, such as drug labels, may follow general structural guidelines but lack further details.
Uncertainty
Biomedical literature contains statements about observations that may not be statements of fact. This text may express uncertainty or skepticism about claims. Without specific adaptations, text mining approaches designed to identify claims within text may mis-characterize these "hedged" statements as facts.
Supporting clinical needs
Biomedical text mining applications developed for clinical use should ideally reflect the needs and demands of clinicians. This is a concern in environments where clinical decision support is expected to be informative and accurate.
Interoperability with clinical systems
New text mining systems must work with existing standards, electronic medical records, and databases. Methods for interfacing with clinical systems such as LOINC have been developed but require extensive organizational effort to implement and maintain.
Patient privacy
Text mining systems operating with private medical data must respect its security and ensure it is rendered anonymous where appropriate.
Processes
Specific sub tasks are of particular concern when processing biomedical text.
Named entity recognition
Developments in biomedical text mining have incorporated identification of biological entities with named entity recognition, or NER. Names and identifiers for biomolecules such as proteins and genes, chemical compounds and drugs, and disease names have all been used as entities. Most entity recognition methods are supported by pre-defined linguistic features or vocabularies, though methods incorporating deep learning and word embeddings have also been successful at biomedical NER.
Document classification and clustering
Biomedical documents may be classified or clustered based on their contents and topics. In classification, document categories are specified manually, while in clustering, documents form algorithm-dependent, distinct groups. These two tasks are representative of supervised and unsupervised methods, respectively, yet the goal of both is to produce subsets of documents based on their distinguishing features. Methods for biomedical document clustering have relied upon k-means clustering.
Relationship discovery
Biomedical documents describe connections between concepts, whether they are interactions between biomolecules, events occurring subsequently over time (i.e., temporal relationships), or causal relationships. Text mining methods may perform relation discovery to identify these connections, often in concert with named entity recognition.
Hedge cue detection
The challenge of identifying uncertain or "hedged" statements has been addressed through hedge cue detection in biomedical literature.
Claim detection
Multiple researchers have developed methods to identify specific scientific claims from literature. In practice, this process involves both isolating phrases and sentences denoting the core arguments made by the authors of a document (a process known as argument mining, employing tools used in fields such as political science) and comparing claims to find potential contradictions between them.
Information extraction
Information extraction, or IE, is the process of automatically identifying structured information from unstructured or partially structured text. IE processes can involve several or all of the above activities, including named entity recognition, relationship discovery, and document classification, with the overall goal of translating text to a more structured form, such as the contents of a template or knowledge base. In the biomedical domain, IE is used to generate links between concepts described in text, such as gene A inhibits gene B and gene C is involved in disease G. Biomedical knowledge bases containing this type of information are generally products of extensive manual curation, so replacement of manual efforts with automated methods remains a compelling area of research.
Information retrieval and question answering
Biomedical text mining supports applications for identifying documents and concepts matching search queries. Search engines such as PubMed search allow users to query literature databases with words or phrases present in document contents, metadata, or indices such as MeSH. Similar approaches may be used for medical literature retrieval. For more fine-grained results, some applications permit users to search with natural language queries and identify specific biomedical relationships.
On 16 March 2020, the National Library of Medicine and others launched the COVID-19 Open Research Dataset (CORD-19) to enable text mining of the current literature on the novel virus. The dataset is hosted by the Semantic Scholar project of the Allen Institute for AI. Other participants include Google, Microsoft Research, the Center for Security and Emerging Technology, and the Chan Zuckerberg Initiative.
Resources
Word embeddings
Several groups have developed sets of biomedical vocabulary mapped to vectors of real numbers, known as word vectors or word embeddings. Sources of pre-trained embeddings specific for biomedical vocabulary are listed in the table below. The majority are results of the word2vec model developed by Mikolov et al or variants of word2vec.
Applications
Text mining applications in the biomedical field include computational approaches to assist with studies in protein docking, protein interactions, and protein-disease associations.
Gene cluster identification
Methods for determining the association of gene clusters obtained by microarray experiments with the biological context provided by the corresponding literature have been developed.
Protein interactions
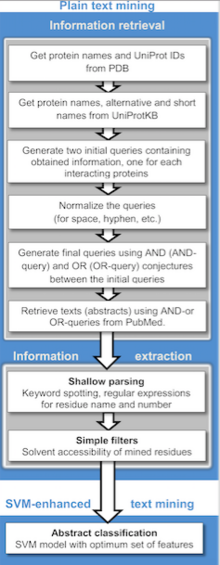
Automatic extraction of protein interactions and associations of proteins to functional concepts (e.g. gene ontology terms) has been explored. The search engine PIE was developed to identify and return protein-protein interaction mentions from MEDLINE-indexed articles. The extraction of kinetic parameters from text or the subcellular location of proteins have also been addressed by information extraction and text mining technology.
Gene-disease associations
Text mining can aid in gene prioritization, or identification of genes most likely to contribute to genetic disease. One group compared several vocabularies, representations and ranking algorithms to develop gene prioritization benchmarks.
Gene-trait associations
An agricultural genomics group identified genes related to bovine reproductive traits using text mining, among other approaches.
Protein-disease associations
Text mining enables an unbiased evaluation of protein-disease relationships within a vast quantity of unstructured textual data.
Applications of phrase mining to disease associations
A text mining study assembled a collection of 709 core extracellular matrix proteins and associated proteins based on two databases: MatrixDB (matrixdb.univ-lyon1.fr) and UniProt. This set of proteins had a manageable size and a rich body of associated information, making it a suitable for the application of text mining tools. The researchers conducted phrase-mining analysis to cross-examine individual extracellular matrix proteins across the biomedical literature concerned with six categories of cardiovascular diseases. They used a phrase-mining pipeline, Context-aware Semantic Online Analytical Processing (CaseOLAP), then semantically scored all 709 proteins according to their Integrity, Popularity, and Distinctiveness using the CaseOLAP pipeline. The text mining study validated existing relationships and informed previously unrecognized biological processes in cardiovascular pathophysiology.
Software tools
Search engines
Search engines designed to retrieve biomedical literature relevant to a user-provided query frequently rely upon text mining approaches. Publicly available tools specific for research literature include PubMed search, Europe PubMed Central search, GeneView, and APSE Similarly, search engines and indexing systems specific for biomedical data have been developed, including DataMed and OmicsDI.
Some search engines, such as Essie, OncoSearch, PubGene, and GoPubMed were previously public but have since been discontinued, rendered obsolete, or integrated into commercial products.
Medical record analysis systems
Electronic medical records (EMRs) and electronic health records (EHRs) are collected by clinical staff in the course of diagnosis and treatment. Though these records generally include structured components with predictable formats and data types, the remainder of the reports are often free-text. Numerous complete systems and tools have been developed to analyse these free-text portions. The MedLEE system was originally developed for analysis of chest radiology reports but later extended to other report topics. The clinical Text Analysis and Knowledge Extraction System, or cTAKES, annotates clinical text using a dictionary of concepts. The CLAMP system offers similar functionality with a user-friendly interface.
Frameworks
Computational frameworks have been developed to rapidly build tools for biomedical text mining tasks. SwellShark is a framework for biomedical NER that requires no human-labeled data but does make use of resources for weak supervision (e.g., UMLS semantic types). The SparkText framework uses Apache Spark data streaming, a NoSQL database, and basic machine learning methods to build predictive models from scientific articles.
APIs
Some biomedical text mining and natural language processing tools are available through application programming interfaces, or APIs. NOBLE Coder performs concept recognition through an API.