From Wikipedia, the free encyclopedia
Cartoon
representation of a proteasome. Its active sites are sheltered inside
the tube (blue). The caps (red; in this case, 11S regulatory particles)
on the ends regulate entry into the destruction chamber, where the
protein is degraded.
Top view of the proteasome above.
Proteasomes are part of a major mechanism by which
cells regulate the
concentration of particular proteins and degrade
misfolded proteins. Proteins are tagged for degradation with a small protein called
ubiquitin. The tagging reaction is catalyzed by enzymes called
ubiquitin ligases.
Once a protein is tagged with a single ubiquitin molecule, this is a
signal to other ligases to attach additional ubiquitin molecules. The
result is a
polyubiquitin chain that is bound by the proteasome, allowing it to degrade the tagged protein. The degradation process yields
peptides of about seven to eight
amino acids long, which can then be further degraded into shorter amino acid sequences and used in
synthesizing new proteins.
In
structure,
the proteasome is a cylindrical complex containing a "core" of four
stacked rings forming a central pore. Each ring is composed of seven
individual proteins. The inner two rings are made of seven
β subunits that contain three to seven protease
active sites.
These sites are located on the interior surface of the rings, so that
the target protein must enter the central pore before it is degraded.
The outer two rings each contain seven
α subunits whose function
is to maintain a "gate" through which proteins enter the barrel. These α
subunits are controlled by binding to "cap" structures or
regulatory particles
that recognize polyubiquitin tags attached to protein substrates and
initiate the degradation process. The overall system of ubiquitination
and proteasomal degradation is known as the
ubiquitin-proteasome system.
Discovery
Before the discovery of the ubiquitin proteasome system, protein degradation in cells was thought to rely mainly on
lysosomes, membrane-bound
organelles with
acidic and
protease-filled interiors that can degrade and then recycle exogenous proteins and aged or damaged organelles. However, work by Joseph Etlinger and
Alfred Goldberg in 1977 on ATP-dependent protein degradation in
reticulocytes, which lack lysosomes, suggested the presence of a second intracellular degradation mechanism. This was shown in 1978 to be composed of several distinct protein chains, a novelty among proteases at the time. Later work on modification of
histones led to the identification of an unexpected
covalent modification of the histone protein by a bond between a
lysine side chain of the histone and the
C-terminal glycine residue of
ubiquitin, a protein that had no known function.
It was then discovered that a previously identified protein associated
with proteolytic degradation, known as ATP-dependent proteolysis factor 1
(APF-1), was the same protein as ubiquitin.
The proteolytic activities of this system were isolated as a
multi-protein complex originally called the multi-catalytic proteinase
complex by Sherwin Wilk and Marion Orlowski. Later, the
ATP-dependent
proteolytic complex that was responsible for ubiquitin-dependent
protein degradation was discovered and was called the 26S proteasome.
Much of the early work leading up to the discovery of the
ubiquitin proteasome system occurred in the late 1970s and early 1980s
at the
Technion in the laboratory of
Avram Hershko, where
Aaron Ciechanover worked as a graduate student. Hershko's year-long sabbatical in the laboratory of
Irwin Rose at the
Fox Chase Cancer Center provided key conceptual insights, though Rose later downplayed his role in the discovery. The three shared the 2004
Nobel Prize in Chemistry for their work in discovering this system.
Although
electron microscopy data revealing the stacked-ring structure of the proteasome became available in the mid-1980s, the first structure of the proteasome core particle was not solved by
X-ray crystallography until 1994.
Structure and organization
A
schematic diagram of the proteasome 20S core particle viewed from one
side. The α subunits that make up the outer two rings are shown in
green, and the β subunits that make up the inner two rings are shown in
blue.
The proteasome subcomponents are often referred to by their
Svedberg sedimentation coefficient (denoted
S). The proteasome most exclusively used in mammals is the cytosolic 26S proteasome, which is about 2000
kilodaltons (kDa) in
molecular mass
containing one 20S protein subunit and two 19S regulatory cap subunits.
The core is hollow and provides an enclosed cavity in which proteins
are degraded; openings at the two ends of the core allow the target
protein to enter. Each end of the core particle associates with a 19S
regulatory subunit that contains multiple
ATPase active sites
and ubiquitin binding sites; it is this structure that recognizes
polyubiquitinated proteins and transfers them to the catalytic core. An
alternative form of regulatory subunit called the 11S particle can
associate with the core in essentially the same manner as the 19S
particle; the 11S may play a role in degradation of foreign peptides
such as those produced after infection by a
virus.
20S core particle
The
number and diversity of subunits contained in the 20S core particle
depends on the organism; the number of distinct and specialized subunits
is larger in multicellular than unicellular organisms and larger in
eukaryotes than in prokaryotes. All 20S particles consist of four
stacked heptameric ring structures that are themselves composed of two
different types of subunits; α subunits are structural in nature,
whereas β subunits are predominantly
catalytic.
The outer two rings in the stack consist of seven α subunits each,
which serve as docking domains for the regulatory particles and the
alpha subunits N-termini form a gate that blocks unregulated access of
substrates to the interior cavity.
The inner two rings each consist of seven β subunits and contain the
protease active sites that perform the proteolysis reactions. Three
distinct catalytic activities were identified in the purified complex:
chymotrypsin-like, trypsin-like and peptidylglutamyl-peptide
hydrolyzing. The size of the proteasome is relatively conserved and is about 150
angstroms
(Å) by 115 Å. The interior chamber is at most 53 Å wide, though the
entrance can be as narrow as 13 Å, suggesting that substrate proteins
must be at least partially unfolded to enter.
In
archaea such as
Thermoplasma acidophilum, all the α and all the β subunits are identical, whereas eukaryotic proteasomes such as those in
yeast contain seven distinct types of each subunit. In
mammals,
the β1, β2, and β5 subunits are catalytic; although they share a common
mechanism, they have three distinct substrate specificities considered
chymotrypsin-like,
trypsin-like, and
peptidyl-glutamyl peptide-hydrolyzing (PHGH). Alternative β forms denoted β1i, β2i, and β5i can be expressed in
hematopoietic cells in response to exposure to pro-
inflammatory signals such as
cytokines, in particular,
interferon gamma. The proteasome assembled with these alternative subunits is known as the
immunoproteasome, whose substrate specificity is altered relative to the normal proteasome.
Recently an alternative proteasome was identified in human cells that lack the α3 core subunit.
These proteasomes (known as the α4-α4 proteasomes) instead form 20S
core particles containing an additional α4 subunit in place of the
missing α3 subunit. These alternative 'α4-α4' proteasomes have been
known previously to exist in yeast.
Although the precise function of these proteasome isoforms is still
largely unknown, cells expressing these proteasomes show enhanced
resistance to toxicity induced by metallic ions such as cadmium.
19S regulatory particle
The
19S particle in eukaryotes consists of 19 individual proteins and is
divisible into two subassemblies, a 9-subunit base that binds directly
to the α ring of the 20S core particle, and a 10-subunit lid. Six of the
nine base proteins are ATPase subunits from the AAA Family, and an
evolutionary homolog of these ATPases exists in archaea, called PAN
(Proteasome-Activating Nucleotidase).
The association of the 19S and 20S particles requires the binding of
ATP to the 19S ATPase subunits, and ATP hydrolysis is required for the
assembled complex to degrade folded and ubiquitinated proteins. Note
that only the step of substrate unfolding requires energy from ATP
hydrolysis, while ATP-binding alone can support all the other steps
required for protein degradation (e.g., complex assembly, gate opening,
translocation, and proteolysis).
In fact, ATP binding to the ATPases by itself supports the rapid
degradation of unfolded proteins. However, while ATP hydrolysis is
required for unfolding only, it is not yet clear whether this energy may
be used in the coupling of some of these steps.
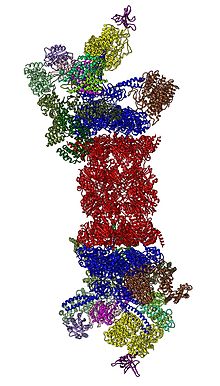
Cartoon representation of the 26S proteasome.
In 2012, two independent efforts have elucidated the molecular architecture of the 26S proteasome by
single particle electron microscopy. More recently, a pseudo-atomic atomic model has been built, again using cryo-EM. In the heart of the 19S, directly adjacent to the 20S, are the AAA-ATPases (
AAA proteins)
that assemble to a heterohexameric ring of the order
Rpt1/Rpt2/Rpt6/Rpt3/Rpt4/Rpt5. This ring is a trimer of dimers:
Rpt1/Rpt2, Rpt6/Rpt3, and Rpt4/Rpt5 dimerize via their N-terminal
coiled-coils. These coiled-coils protrude from the hexameric ring. The
largest regulatory particle non-ATPases Rpn1 and Rpn2 bind to the tips
of Rpt1/2 and Rpt6/3, respectively. The ubiquitin receptor Rpn13 binds
to Rpn2 and completes the base cub-complex. The lid covers one half of
the AAA-ATPase hexamer (Rpt6/Rpt3/Rpt4) and, unexpectedly, directly
contacts the 20S via Rpn6 and to lesser extent Rpn5. The subunits Rpn9,
Rpn5, Rpn6, Rpn7, Rpn3, and Rpn12, which are structurally related among
themselves and to subunits of the
COP9 complex and
eIF3
(hence called PCI subunits) assemble to a horseshoe-like structure
enclosing the Rpn8/Rpn11 heterodimer. Rpn11, the deubiquinating enzyme,
is placed at the mouth of the AAA-ATPase hexamer, ideally positioned to
remove ubiquitin moieties immediately before translocation of substrates
into the 20S. The second ubiquitin receptor identified to date, Rpn10,
is positioned at the periphery of the lid, near subunits Rpn8 and Rpn9.
Conformational changes of 19S
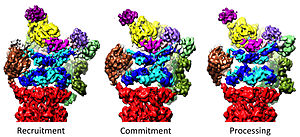
The 19S regulatory particle has been observed in three strongly differing conformational states to date.
Realization of all these three conformational states is likely
necessary for accomplishing substrate recognition and degradation (see
below). A hallmark of the AAA-ATPase configuration in this predominant
low-energy state is a staircase- or lockwasher-like arrangement of the
AAA-domains. Also in the presence of
ATP
but absence of substrate an alternative, less abundant conformation of
the 19S is adopted primarily differing in the positioning of the lid
with respect to the AAA-ATPase module.
In the presence of ATP-gammaS or a substrate (stabilized in a 26S
mutant with defective Rpn11) a third conformation has been observed
displaying a dramatic structural change of the AAA-ATPase module.
Three distinct conformational states of the 26S proteasome.
The conformations are hypothesized to be responsible for recruitment of
the substrate, its irreversible commitment, and finally processing and
translocation into the core particle, where degradation occurs.
Regulation of the 20S by the 19S
The
19S regulatory particle is responsible for stimulating the 20S to
degrade proteins. A primary function of the 19S regulatory ATPases is to
open the gate in the 20S that blocks the entry of substrates into the
degradation chamber. The mechanism by which the proteasomal ATPase open this gate has been recently elucidated. 20S gate opening, and thus substrate degradation, requires the C-termini of the proteasomal ATPases, which contains a specific
motif
(i.e., HbYX motif). The ATPases C-termini bind into pockets in the top
of the 20S, and tether the ATPase complex to the 20S proteolytic
complex, thus joining the substrate unfolding equipment with the 20S
degradation machinery. Binding of these C-termini into these 20S pockets
by themselves stimulates opening of the gate in the 20S in much the
same way that a "key-in-a-lock" opens a door. The precise mechanism by which this "key-in-a-lock" mechanism functions has been structurally elucidated.
11S regulatory particle
20S
proteasomes can also associate with a second type of regulatory
particle, the 11S regulatory particle, a heptameric structure that does
not contain any ATPases and can promote the degradation of short
peptides
but not of complete proteins. It is presumed that this is because the
complex cannot unfold larger substrates. This structure is also known as
PA28 or REG. The mechanisms by which it binds to the core particle
through the C-terminal tails of its subunits and induces α-ring
conformational changes to open the 20S gate suggest a similar mechanism for the 19S particle.
The expression of the 11S particle is induced by interferon gamma and
is responsible, in conjunction with the immunoproteasome β subunits, for
the generation of peptides that bind to the
major histocompatibility complex.
Assembly
The
assembly of the proteasome is a complex process due to the number of
subunits that must associate to form an active complex. The β subunits
are synthesized with
N-terminal "propeptides" that are
post-translationally modified
during the assembly of the 20S particle to expose the proteolytic
active site. The 20S particle is assembled from two half-proteasomes,
each of which consists of a seven-membered pro-β ring attached to a
seven-membered α ring. The association of the β rings of the two
half-proteasomes triggers
threonine-dependent
autolysis of the propeptides to expose the active site. These β interactions are mediated mainly by
salt bridges and
hydrophobic interactions between conserved
alpha helices whose disruption by
mutation damages the proteasome's ability to assemble.
The assembly of the half-proteasomes, in turn, is initiated by the
assembly of the α subunits into their heptameric ring, forming a
template for the association of the corresponding pro-β ring. The
assembly of α subunits has not been characterized.
Only recently, the assembly process of the 19S regulatory
particle has been elucidated to considerable extent. The 19S regulatory
particle assembles as two distinct subcomponents, the base and the lid.
Assembly of the base complex is facilitated by four assembly
chaperones, Hsm3/S5b, Nas2/p27, Rpn14/PAAF1, and Nas6/
gankyrin (names for yeast/mammals). These assembly chaperones bind to the AAA-
ATPase subunits and their main function seems to be to ensure proper assembly of the heterohexameric AAA-
ATPase
ring. To date it is still under debate whether the base complex
assembles separately, whether the assembly is templated by the 20S core
particle, or whether alternative assembly pathways exist. In addition to
the four assembly chaperones, the deubiquitinating enzyme Ubp6/
Usp14 also promotes base assembly, but it is not essential. The lid assembles separately in a specific order and does not require assembly chaperones.
The protein degradation process
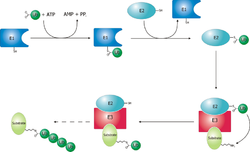
Ubiquitination and targeting
Proteins are targeted for degradation by the proteasome with covalent
modification of a lysine residue that requires the coordinated
reactions of three
enzymes. In the first step, a
ubiquitin-activating enzyme (known as E1) hydrolyzes ATP and adenylylates a
ubiquitin molecule. This is then transferred to E1's active-site
cysteine residue in concert with the adenylylation of a second ubiquitin. This adenylylated ubiquitin is then transferred to a cysteine of a second enzyme,
ubiquitin-conjugating enzyme (E2). In the last step, a member of a highly diverse class of enzymes known as
ubiquitin ligases
(E3) recognizes the specific protein to be ubiquitinated and catalyzes
the transfer of ubiquitin from E2 to this target protein. A target
protein must be labeled with at least four ubiquitin monomers (in the
form of a polyubiquitin chain) before it is recognized by the proteasome
lid. It is therefore the E3 that confers
substrate specificity to this system.
The number of E1, E2, and E3 proteins expressed depends on the organism
and cell type, but there are many different E3 enzymes present in
humans, indicating that there is a huge number of targets for the
ubiquitin proteasome system.
The mechanism by which a polyubiquitinated protein is targeted to
the proteasome is not fully understood. Ubiquitin-receptor proteins
have an
N-terminal
ubiquitin-like (UBL) domain and one or more ubiquitin-associated (UBA)
domains. The UBL domains are recognized by the 19S proteasome caps and
the UBA domains bind ubiquitin via
three-helix bundles.
These receptor proteins may escort polyubiquitinated proteins to the
proteasome, though the specifics of this interaction and its regulation
are unclear.
The
ubiquitin protein itself is 76
amino acids long and was named due to its ubiquitous nature, as it has a highly
conserved sequence and is found in all known eukaryotic organisms. The genes encoding ubiquitin in
eukaryotes are arranged in
tandem repeats, possibly due to the heavy
transcription demands on these genes to produce enough ubiquitin for the cell. It has been proposed that ubiquitin is the slowest-
evolving protein identified to date.
Ubiquitin contains seven lysine residues to which another ubiquitin can
be ligated, resulting in different types of polyubiquitin chains.
Chains in which each additional ubiquitin is linked to lysine 48 of the
previous ubiquitin have a role in proteasome targeting, while other
types of chains may be involved in other processes.
Unfolding and translocation
After a protein has been ubiquitinated, it is recognized by the 19S regulatory particle in an ATP-dependent binding step.
The substrate protein must then enter the interior of the 20S particle
to come in contact with the proteolytic active sites. Because the 20S
particle's central channel is narrow and gated by the N-terminal tails
of the α ring subunits, the substrates must be at least partially
unfolded before they enter the core. The passage of the unfolded
substrate into the core is called
translocation and necessarily occurs after deubiquitination. However, the order in which substrates are deubiquitinated and unfolded is not yet clear. Which of these processes is the
rate-limiting step
in the overall proteolysis reaction depends on the specific substrate;
for some proteins, the unfolding process is rate-limiting, while
deubiquitination is the slowest step for other proteins. The extent to which substrates must be unfolded before translocation is not known, but substantial
tertiary structure, and in particular nonlocal interactions such as
disulfide bonds, are sufficient to inhibit degradation. The presence of
intrinsically disordered protein
segments of sufficient size, either at the protein terminus or
internally, has also been proposed to facilitate efficient initiation of
degradation.
The gate formed by the α subunits prevents peptides longer than
about four residues from entering the interior of the 20S particle. The
ATP molecules bound before the initial recognition step are
hydrolyzed before translocation. While energy is needed for substrate unfolding, it is not required for translocation. The assembled 26S proteasome can degrade unfolded proteins in the presence of a non-hydrolyzable
ATP analog, but cannot degrade folded proteins, indicating that energy from ATP hydrolysis is used for substrate unfolding. Passage of the unfolded substrate through the opened gate occurs via
facilitated diffusion if the 19S cap is in the ATP-bound state.
The mechanism for unfolding of
globular proteins is necessarily general, but somewhat dependent on the
amino acid sequence. Long sequences of alternating glycine and
alanine
have been shown to inhibit substrate unfolding, decreasing the
efficiency of proteasomal degradation; this results in the release of
partially degraded byproducts, possibly due to the decoupling of the ATP
hydrolysis and unfolding steps. Such glycine-alanine repeats are also found in nature, for example in
silk fibroin; in particular, certain
Epstein–Barr virus gene products bearing this sequence can stall the proteasome, helping the virus propagate by preventing
antigen presentation on the major histocompatibility complex.
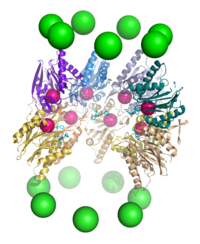
A cutaway view of the proteasome 20S core particle illustrating the locations of the active sites. The α subunits are represented as green spheres and the β subunits as protein backbones colored by individual polypeptide chain. The small pink spheres represent the location of the active-site threonine residue in each subunit. Light blue chemical structures are the inhibitor bortezomib bound to the active sites.
Proteolysis
The mechanism of proteolysis by the β subunits of the 20S core particle is through a threonine-dependent
nucleophilic attack. This mechanism may depend on an associated
water molecule for deprotonation of the reactive threonine
hydroxyl.
Degradation occurs within the central chamber formed by the association
of the two β rings and normally does not release partially degraded
products, instead reducing the substrate to short polypeptides typically
7–9 residues long, though they can range from 4 to 25 residues,
depending on the organism and substrate. The biochemical mechanism that
determines product length is not fully characterized.
Although the three catalytic β subunits have a common mechanism, they
have slightly different substrate specificities, which are considered
chymotrypsin-like, trypsin-like, and peptidyl-glutamyl
peptide-hydrolyzing (PHGH)-like. These variations in specificity are the
result of interatomic contacts with local residues near the active
sites of each subunit. Each catalytic β subunit also possesses a
conserved lysine residue required for proteolysis.
Although the proteasome normally produces very short peptide
fragments, in some cases these products are themselves biologically
active and functional molecules. Certain
transcription factors regulating the expression of specific genes, including one component of the mammalian complex
NF-κB,
are synthesized as inactive precursors whose ubiquitination and
subsequent proteasomal degradation converts them to an active form. Such
activity requires the proteasome to cleave the substrate protein
internally, rather than processively degrading it from one terminus. It
has been suggested that long
loops
on these proteins' surfaces serve as the proteasomal substrates and
enter the central cavity, while the majority of the protein remains
outside. Similar effects have been observed in yeast proteins; this mechanism of selective degradation is known as
regulated ubiquitin/proteasome dependent processing (RUP).
Ubiquitin-independent degradation
Although
most proteasomal substrates must be ubiquitinated before being
degraded, there are some exceptions to this general rule, especially
when the proteasome plays a normal role in the post-
translational processing of the protein. The proteasomal activation of NF-κB by processing
p105 into p50 via internal proteolysis is one major example. Some proteins that are hypothesized to be unstable due to
intrinsically unstructured regions,
are degraded in a ubiquitin-independent manner. The most well-known
example of a ubiquitin-independent proteasome substrate is the enzyme
ornithine decarboxylase. Ubiquitin-independent mechanisms targeting key
cell cycle regulators such as
p53 have also been reported, although p53 is also subject to ubiquitin-dependent degradation.
Finally, structurally abnormal, misfolded, or highly oxidized proteins
are also subject to ubiquitin-independent and 19S-independent
degradation under conditions of cellular stress.
Evolution
The 20S proteasome is both ubiquitous and essential in eukaryotes. Some
prokaryotes, including many archaea and the
bacterial order
Actinomycetales, also share homologs of the 20S proteasome, whereas most bacteria possess
heat shock genes
hslV and
hslU, whose gene products are a multimeric protease arranged in a two-layered ring and an ATPase. The hslV protein has been hypothesized to resemble the likely ancestor of the 20S proteasome. In general, HslV is not essential in bacteria, and not all bacteria possess it, whereas some
protists possess both the 20S and the hslV systems.
Many bacteria also possess other homologs of the proteasome and an
associated ATPase, most notably ClpP and ClpX. This redundancy explains
why the HslUV system is not essential.
Sequence analysis suggests that the catalytic β subunits diverged
earlier in evolution than the predominantly structural α subunits. In
bacteria that express a 20S proteasome, the β subunits have high
sequence identity
to archaeal and eukaryotic β subunits, whereas the α sequence identity
is much lower. The presence of 20S proteasomes in bacteria may result
from
lateral gene transfer, while the diversification of subunits among eukaryotes is ascribed to multiple
gene duplication events.
Cell cycle control
Cell cycle progression is controlled by ordered action of
cyclin-dependent kinases (CDKs), activated by specific
cyclins that demarcate phases of the
cell cycle.
Mitotic cyclins, which persist in the cell for only a few minutes, have
one of the shortest life spans of all intracellular proteins.
After a CDK-cyclin complex has performed its function, the associated
cyclin is polyubiquitinated and destroyed by the proteasome, which
provides directionality for the cell cycle. In particular, exit from
mitosis requires the proteasome-dependent dissociation of the regulatory component
cyclin B from the
mitosis promoting factor complex. In
vertebrate cells, "slippage" through the mitotic checkpoint leading to premature
M phase exit can occur despite the delay of this exit by the
spindle checkpoint.
Earlier cell cycle checkpoints such as post-
restriction point check between
G1 phase and
S phase similarly involve proteasomal degradation of
cyclin A, whose ubiquitination is promoted by the
anaphase promoting complex (APC), an E3
ubiquitin ligase. The APC and the Skp1/Cul1/F-box protein complex (
SCF complex)
are the two key regulators of cyclin degradation and checkpoint
control; the SCF itself is regulated by the APC via ubiquitination of
the adaptor protein, Skp2, which prevents SCF activity before the G1-S
transition.
Individual components of the 19S particle have their own regulatory roles.
Gankyrin, a recently identified
oncoprotein, is one of the 19S subcomponents that also tightly binds the
cyclin-dependent kinase CDK4 and plays a key role in recognizing ubiquitinated
p53, via its affinity for the ubiquitin ligase
MDM2. Gankyrin is anti-
apoptotic and has been shown to be overexpressed in some
tumor cell types such as
hepatocellular carcinoma.
Regulation of plant growth
In
plants, signaling by
auxins, or
phytohormones that order the direction and
tropism of plant growth, induces the targeting of a class of
transcription factor
repressors known as Aux/IAA proteins for proteasomal degradation. These
proteins are ubiquitinated by SCFTIR1, or SCF in complex with the auxin
receptor TIR1. Degradation of Aux/IAA proteins derepresses
transcription factors in the auxin-response factor (ARF) family and
induces ARF-directed gene expression.
The cellular consequences of ARF activation depend on the plant type
and developmental stage, but are involved in directing growth in roots
and leaf veins. The specific response to ARF derepression is thought to
be mediated by specificity in the pairing of individual ARF and Aux/IAA
proteins.
Apoptosis
Both internal and external
signals can lead to the induction of
apoptosis,
or programmed cell death. The resulting deconstruction of cellular
components is primarily carried out by specialized proteases known as
caspases,
but the proteasome also plays important and diverse roles in the
apoptotic process. The involvement of the proteasome in this process is
indicated by both the increase in protein ubiquitination, and of E1, E2,
and E3 enzymes that is observed well in advance of apoptosis. During apoptosis, proteasomes localized to the nucleus have also been observed to translocate to outer membrane
blebs characteristic of apoptosis.
Proteasome inhibition has different effects on apoptosis
induction in different cell types. In general, the proteasome is not
required for apoptosis, although inhibiting it is pro-apoptotic in most
cell types that have been studied. Apoptosis is mediated through
disrupting the regulated degradation of pro-growth cell cycle proteins. However, some cell lines — in particular,
primary cultures of
quiescent and
differentiated cells such as
thymocytes and
neurons —
are prevented from undergoing apoptosis on exposure to proteasome
inhibitors. The mechanism for this effect is not clear, but is
hypothesized to be specific to cells in quiescent states, or to result
from the differential activity of the pro-apoptotic
kinase JNK.
The ability of proteasome inhibitors to induce apoptosis in rapidly
dividing cells has been exploited in several recently developed
chemotherapy agents such as
bortezomib and
salinosporamide A.
Response to cellular stress
In response to cellular stresses – such as
infection,
heat shock, or
oxidative damage –
heat shock proteins that identify misfolded or unfolded proteins and target them for proteasomal degradation are expressed. Both
Hsp27 and
Hsp90—
chaperone
proteins have been implicated in increasing the activity of the
ubiquitin-proteasome system, though they are not direct participants in
the process.
Hsp70, on the other hand, binds exposed
hydrophobic
patches on the surface of misfolded proteins and recruits E3 ubiquitin
ligases such as CHIP to tag the proteins for proteasomal degradation.
The CHIP protein (carboxyl terminus of Hsp70-interacting protein) is
itself regulated via inhibition of interactions between the E3 enzyme
CHIP and its E2 binding partner.
Similar mechanisms exist to promote the degradation of
oxidatively damaged proteins via the proteasome system. In particular, proteasomes localized to the nucleus are regulated by
PARP and actively degrade inappropriately oxidized
histones.
Oxidized proteins, which often form large amorphous aggregates in the
cell, can be degraded directly by the 20S core particle without the 19S
regulatory cap and do not require ATP hydrolysis or tagging with
ubiquitin.
However, high levels of oxidative damage increases the degree of
cross-linking between protein fragments, rendering the aggregates
resistant to proteolysis. Larger numbers and sizes of such highly
oxidized aggregates are associated with
aging.
Dysregulation of the ubiquitin proteasome system may contribute to several neural diseases. It may lead to brain tumors such as
astrocytomas. In some of the late-onset
neurodegenerative diseases that share aggregation of misfolded proteins as a common feature, such as
Parkinson's disease and
Alzheimer's disease, large insoluble aggregates of misfolded proteins can form and then result in
neurotoxicity,
through mechanisms that are not yet well understood. Decreased
proteasome activity has been suggested as a cause of aggregation and
Lewy body formation in Parkinson's. This hypothesis is supported by the observation that
yeast models of Parkinson's are more susceptible to toxicity from
α-synuclein, the major protein component of Lewy bodies, under conditions of low proteasome activity. Impaired proteasomal activity may underlie cognitive disorders such as the
autism spectrum disorders, and muscle and nerve diseases such as
inclusion body myopathy.
Role in the immune system
The proteasome plays a straightforward but critical role in the function of the
adaptive immune system. Peptide
antigens are displayed by the
major histocompatibility complex class I (MHC) proteins on the surface of
antigen-presenting cells. These peptides are products of proteasomal degradation of proteins originated by the invading
pathogen.
Although constitutively expressed proteasomes can participate in this
process, a specialized complex composed of proteins, whose
expression is induced by
interferon gamma,
are the primary producers of peptides which are optimal in size and
composition for MHC binding. These proteins whose expression increases
during the immune response include the 11S regulatory particle, whose
main known biological role is regulating the production of MHC ligands,
and specialized β subunits called β1i, β2i, and β5i with altered
substrate specificity. The complex formed with the specialized β
subunits is known as the
immunoproteasome.
Another β5i variant subunit, β5t, is expressed in the thymus, leading
to a thymus-specific "thymoproteasome" whose function is as yet unclear.
The strength of MHC class I ligand binding is dependent on the composition of the ligand
C-terminus, as peptides bind by
hydrogen bonding
and by close contacts with a region called the "B pocket" on the MHC
surface. Many MHC class I alleles prefer hydrophobic C-terminal
residues, and the immunoproteasome complex is more likely to generate
hydrophobic C-termini.
Proteasome inhibitors
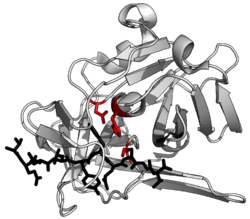
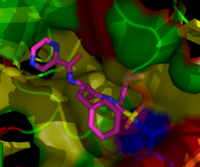
Bortezomib bound to the core particle in a yeast proteasome. The bortezomib molecule is in the center colored by atom type (carbon = pink, nitrogen = blue, oxygen = red, boron = yellow), surrounded by the local protein surface. The blue patch is the catalytic threonine residue whose activity is blocked by the presence of bortezomib.
Proteasome inhibitors have effective anti-
tumor activity in
cell culture, inducing
apoptosis by disrupting the regulated degradation of pro-growth cell cycle proteins. This approach of selectively inducing apoptosis in tumor cells has proven effective in animal models and human trials.
Lactacystin, a natural product synthesized by
Streptomyces bacteria, was the first non-peptidic proteasome inhibitor discovered
and is widely used as a research tool in biochemistry and cell biology.
Lactacystin was licensed to Myogenics/Proscript, which was acquired by
Millennium Pharmaceuticals, now part of
Takeda Pharmaceuticals.
Lactacystin covalently modifies the amino-terminal threonine of
catalytic β subunits of the proteasome, particularly the β5 subunit
responsible for the proteasome's chymotrypsin-like activity. This
discovery helped to establish the proteasome as a mechanistically novel
class of protease: an amino-terminal
threonine protease.
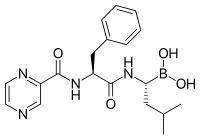
Bortezomib (Boronated MG132), a molecule developed by
Millennium Pharmaceuticals and marketed as Velcade, is the first proteasome inhibitor to reach clinical use as a
chemotherapy agent. Bortezomib is used in the treatment of
multiple myeloma. Notably, multiple myeloma has been observed to result in increased proteasome-derived peptide levels in
blood serum that decrease to normal levels in response to successful chemotherapy. Studies in animals have indicated that bortezomib may also have clinically significant effects in
pancreatic cancer. Preclinical and early clinical studies have been started to examine bortezomib's effectiveness in treating other
B-cell-related cancers, particularly some types of
non-Hodgkin's lymphoma.
Clinical results also seem to justify use of proteasome inhibitor
combined with chemotherapy, for B-cell acute lymphoblastic leukemia Proteasome inhibitors can kill some types of cultured leukemia cells that are resistant to glucocorticoids.
The molecule
ritonavir, marketed as Norvir, was developed as a
protease inhibitor and used to target
HIV infection. However, it has been shown to inhibit proteasomes as well as free proteases; to be specific, the
chymotrypsin-like activity of the proteasome is inhibited by ritonavir, while the
trypsin-like activity is somewhat enhanced. Studies in animal models suggest that ritonavir may have inhibitory effects on the growth of
glioma cells.
Proteasome inhibitors have also shown promise in treating
autoimmune diseases in animal models. For example, studies in mice
bearing human
skin grafts found a reduction in the size of lesions from
psoriasis after treatment with a proteasome inhibitor. Inhibitors also show positive effects in
rodent models of
asthma.
Labeling and inhibition of the proteasome is also of interest in laboratory settings for both
in vitro and
in vivo study of proteasomal activity in cells. The most commonly used laboratory inhibitors are
lactacystin and the peptide aldehyde
MG132 initially developed by Goldberg lab.
Fluorescent inhibitors have also been developed to specifically label the active sites of the assembled proteasome.
Clinical significance
The
proteasome and its subunits are of clinical significance for at least
two reasons: (1) a compromised complex assembly or a dysfunctional
proteasome can be associated with the underlying pathophysiology of
specific diseases, and (2) they can be exploited as drug targets for
therapeutic interventions. More recently, more effort has been made to
consider the proteasome for the development of novel diagnostic markers
and strategies. An improved and comprehensive understanding of the
pathophysiology of the proteasome should lead to clinical applications
in the future.
The proteasomes form a pivotal component for the Ubiquitin-Proteasome System (UPS) and corresponding cellular Protein Quality Control (PQC). Protein
ubiquitination and subsequent
proteolysis and degradation by the proteasome are important mechanisms in the regulation of the
cell cycle,
cell growth and differentiation, gene transcription, signal transduction and
apoptosis.
Subsequently, a compromised proteasome complex assembly and function
lead to reduced proteolytic activities and the accumulation of damaged
or misfolded protein species. Such protein accumulation may contribute
to the pathogenesis and phenotypic characteristics in neurodegenerative
diseases, cardiovascular diseases, inflammatory responses and autoimmune diseases, and systemic DNA damage responses leading to
malignancies.
Several experimental and clinical studies have indicated that
aberrations and deregulations of the UPS contribute to the pathogenesis
of several neurodegenerative and myodegenerative disorders, including
Alzheimer's disease,
Parkinson's disease and
Pick's disease,
Amyotrophic lateral sclerosis (
ALS),
Huntington's disease,
Creutzfeldt–Jakob disease, and motor neuron diseases, polyglutamine (PolyQ) diseases,
Muscular dystrophies and several rare forms of neurodegenerative diseases associated with
dementia.
As part of the Ubiquitin-Proteasome System (UPS), the proteasome
maintains cardiac protein homeostasis and thus plays a significant role
in cardiac
Ischemic injury,
ventricular hypertrophy and
Heart failure.
Additionally, evidence is accumulating that the UPS plays an essential
role in malignant transformation. UPS proteolysis plays a major role in
responses of cancer cells to stimulatory signals that are critical for
the development of cancer. Accordingly, gene expression by degradation
of
transcription factors, such as
p53,
c-Jun,
c-Fos,
NF-κB,
c-Myc, HIF-1α, MATα2,
STAT3, sterol-regulated element-binding proteins and
androgen receptors are all controlled by the UPS and thus involved in the development of various malignancies. Moreover, the UPS regulates the degradation of tumor suppressor gene products such as
adenomatous polyposis coli (
APC) in colorectal cancer,
retinoblastoma (Rb). and
von Hippel-Lindau tumor suppressor (VHL), as well as a number of
proto-oncogenes (
Raf,
Myc,
Myb,
Rel,
Src,
Mos,
Abl).
The UPS is also involved in the regulation of inflammatory responses.
This activity is usually attributed to the role of proteasomes in the
activation of NF-κB which further regulates the expression of pro
inflammatory
cytokines such as
TNF-α, IL-β,
IL-8,
adhesion molecules (
ICAM-1,
VCAM-1,
P-selectin) and
prostaglandins and
nitric oxide (NO).
Additionally, the UPS also plays a role in inflammatory responses as
regulators of leukocyte proliferation, mainly through proteolysis of
cyclines and the degradation of
CDK inhibitors. Lastly,
autoimmune disease patients with
SLE,
Sjogren's syndrome and
rheumatoid arthritis (RA) predominantly exhibit circulating proteasomes which can be applied as clinical biomarkers.