| Severe acute respiratory syndrome coronavirus 2 | |
|---|---|

| |
| Electron micrograph of SARS-CoV-2 virions with visible coronae | |

| |
| Illustration of a SARS-CoV-2 virion | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Phylum: | incertae sedis |
| Order: | Nidovirales |
| Family: | Coronaviridae |
| Genus: | Betacoronavirus |
| Subgenus: | Sarbecovirus |
| Species: | |
| Strain: |
Severe acute respiratory syndrome coronavirus 2
|
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), previously known by the provisional name 2019 novel coronavirus (2019-nCoV), is a positive-sense single-stranded RNA virus. It is contagious in humans and is the cause of the ongoing 2019–20 coronavirus outbreak, an epidemic of coronavirus disease 2019 (COVID-19) that has been designated a Public Health Emergency of International Concern by the World Health Organization (WHO).
SARS-CoV-2 has close genetic similarity to bat coronaviruses, from which it likely originated. An intermediate reservoir such as a pangolin is also thought to be involved in its introduction to humans. From a taxonomic perspective SARS-CoV-2 is classified as a strain of the species severe acute respiratory syndrome-related coronavirus (SARSr-CoV).
Because the strain was first discovered in Wuhan, China, it is sometimes referred to informally as the Wuhan coronavirus, although the World Health Organization (WHO) discourages the use of names based upon locations. To avoid confusion with the disease SARS, the WHO sometimes refers to the virus as "the virus responsible for COVID-19" in public health communications.
Virology
Infection
Human-to-human transmission of SARS-CoV-2 has been confirmed during the 2019–20 coronavirus outbreak. Transmission occurs primarily via respiratory droplets from coughs and sneezes within a range of about 6 feet (1.8 m). Indirect contact via contaminated surfaces is another possible cause of infection. Viral RNA has also been found in stool samples from infected patients.
It is possible that the virus can be infectious even during the incubation period, but this has not been proven, and the World Health Organization
(WHO) stated on 1 February 2020 that "transmission from asymptomatic
cases is likely not a major driver of transmission" at this time. Thus, most infections in humans are believed to be the result of transmission from subjects exhibiting symptoms of coronavirus disease 2019.
Reservoir
The first known infections from the SARS-CoV-2 strain were discovered in Wuhan, China. The original source of viral transmission to humans remains unclear. However, research into the origin of the 2003 SARS outbreak has resulted in the discovery of many SARS-like bat coronaviruses, most originating in the Rhinolophus genus of horseshoe bats. Two viral nucleic acid sequences found in samples taken from Rhinolophus sinicus show a resemblance of 80% to SARS-CoV-2. A third viral nucleic acid sequence from Rhinolophus affinis collected in Yunnan province has a 96% resemblance to SARS-CoV-2. The WHO considers bats the most likely natural reservoir of SARS-CoV-2.
A metagenomic study published in 2019 previously revealed that SARS-CoV, the strain of the virus that causes SARS, was the most widely distributed coronavirus among a sample of Malayan pangolins. On 7 February 2020, it was announced that researchers from Guangzhou had discovered a pangolin sample with a viral nucleic acid sequence "99% identical" to SARS-CoV-2.
When released, the results clarified that "the receptor-binding domain
of the S protein of the newly discovered Pangolin-CoV is virtually
identical to that of 2019-nCoV, with one amino acid difference." Pangolins are protected under Chinese law, but their poaching and trading for use in traditional Chinese medicine remains common.
Microbiologists and geneticists in Texas have independently found evidence of reassortment in coronaviruses suggesting the involvement of pangolins in the origin of SARS-CoV-2. They acknowledged remaining unknown factors while urging continued examination of other mammals.
Phylogenetics and taxonomy
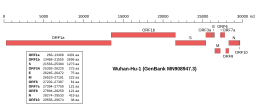
Genomic organisation of isolate Wuhan-Hu-1, the earliest sequenced sample of SARS-CoV-2
| |
| NCBI genome ID | MN908947 |
|---|---|
| Genome size | 29,903 bases |
| Year of completion | 2020 |
SARS-CoV-2 belongs to the broad family of viruses known as coronaviruses. It is a positive-sense single-stranded RNA (+ssRNA) virus. Other coronaviruses are capable of causing illnesses ranging from the common cold to more severe diseases such as Middle East respiratory syndrome (MERS). It is the seventh known coronavirus to infect people, after 229E, NL63, OC43, HKU1, MERS-CoV, and the original SARS-CoV.
Like the SARS-related coronavirus strain implicated in the 2003 SARS outbreak, SARS-CoV-2 is a member of the subgenus Sarbecovirus (beta-CoV lineage B). Its RNA sequence is approximately 30,000 bases in length.
SARS-CoV-2 is unique among known betacoronaviruses in its incorporation
of a polybasic cleavage site, a characteristic known to increase pathogenicity and transmissibility in other viruses.
With a sufficient number of sequenced genomes, it is possible to reconstruct a phylogenetic tree
of the mutation history of a family of viruses. By 12 January 2020,
five genomes of SARS-CoV-2 had been isolated from Wuhan and reported by
the Chinese Center for Disease Control and Prevention (CCDC) and other institutions; the number of genomes increased to 81 by 11 February 2020. A phylogenetic analysis of those samples showed they were "highly related with at most seven mutations relative to a common ancestor", implying that the first human infection occurred in November or December 2019.
On 11 February 2020, the International Committee on Taxonomy of Viruses
(ICTV) announced that according to existing rules that compute
hierarchical relationships among coronaviruses on the basis of five conserved sequences of nucleic acids, the differences between what was then called 2019-nCoV and the virus strain from the 2003 SARS outbreak were insufficient to make it a separate viral species. Therefore, they identified 2019-nCoV as a strain of severe acute respiratory syndrome-related coronavirus.
Structural biology
Digitally colourized electron micrographs of SARS-CoV-2 (yellow) emerging from human cells
Each SARS-CoV-2 virion is approximately 50–200 nanometres in diameter. Like other coronaviruses, SARS-CoV-2 has four structural proteins, known as the S (spike), E (envelope), M (membrane), and N (nucleocapsid) proteins; the N protein holds the RNA genome, and the S, E, and M proteins together create the viral envelope. The spike protein is responsible for allowing the virus to attach to the membrane of a host cell.
Protein modeling experiments on the spike protein of the virus soon suggested that SARS-CoV-2 has sufficient affinity to the angiotensin converting enzyme 2 (ACE2) receptors of human cells to use them as a mechanism of cell entry. By 22 January 2020, a group in China working with the full virus genome and a group in the United States using reverse genetics methods independently and experimentally demonstrated that ACE2 could act as the receptor for SARS-CoV-2. Studies have shown that SARS-CoV-2 has a higher affinity to human ACE2 than the original SARS virus strain. An atomic-level image of the S protein has been created using cryogenic electron microscopy.
SARS-Cov-2 produces at least three virulence factors that promote dissemination of new virions from host cells and inhibit immune response.
Epidemiology
Based upon the low variability exhibited among known SARS-CoV-2 genomic
sequences, the strain is thought to have been detected by health
authorities within weeks of its emergence among the human population in
late 2019.
The virus subsequently spread to all provinces of China and to more
than one hundred other countries in Asia, Europe, North America, South
America, Africa, and Oceania. Human-to-human transmission of the virus has been confirmed in all of these regions. On 30 January 2020, SARS-CoV-2 was designated a Public Health Emergency of International Concern by the WHO.
As of 9 March 2020 (22:40 UTC), there were 113,584 confirmed cases of infection, of which 80,735
were within mainland China. While the proportion of infections that
result in confirmed infection or progress to diagnosable disease remains
unclear,
one mathematical model estimated the number of people infected in Wuhan
alone at 75,815 as of 25 January 2020, at a time when confirmed
infections were far lower. The total number of deaths attributed to the virus was 3,996 as of 9 March 2020 (22:40 UTC). Over 75% of all deaths have occurred in Hubei province, where Wuhan is located.
The basic reproduction number () of the virus has been estimated to be between 1.4 and 3.9.
This means that each infection from the virus is expected to result in
1.4 to 3.9 new infections when no preventive measures are taken.



