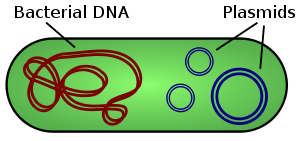
Illustration of a bacterium showing chromosomal DNA and plasmids. Not to scale.
A plasmid is a small DNA molecule within a cell that is physically separated from a chromosomal DNA and can replicate independently. They are most commonly found as small circular, double-stranded DNA molecules in bacteria; however, plasmids are sometimes present in archaea and eukaryotic organisms. In nature, plasmids often carry genes that may benefit the survival of the organism, for example antibiotic resistance.
While the chromosomes are big and contain all the essential genetic
information for living under normal conditions, plasmids usually are
very small and contain only additional genes that may be useful to the
organism under certain situations or particular conditions. Artificial
plasmids are widely used as vectors in molecular cloning, serving to drive the replication of recombinant DNA sequences within host organisms. In the laboratory, plasmids may be introduced into a cell via transformation.
Plasmids are considered replicons, units of DNA capable of replicating autonomously within a suitable host. However, plasmids, like viruses, are not generally classified as life. Plasmids are transmitted from one bacterium to another (even of another species) mostly through conjugation. This host-to-host transfer of genetic material is one mechanism of horizontal gene transfer, and plasmids are considered part of the mobilome. Unlike viruses (which encase their genetic material in a protective protein coat called a capsid),
plasmids are "naked" DNA and do not encode genes necessary to encase
the genetic material for transfer to a new host. However, some classes
of plasmids encode the conjugative "sex" pilus necessary for their own transfer. The size of the plasmid varies from 1 to over 200 kbp, and the number of identical plasmids in a single cell can range anywhere from one to thousands under some circumstances.
The relationship between microbes and plasmid DNA is neither
parasitic nor mutualistic, because each implies the presence of an
independent species living in a detrimental or commensal state with the
host organism. Rather, plasmids provide a mechanism for horizontal gene
transfer within a population of microbes and typically provide a
selective advantage under a given environmental state. Plasmids may
carry genes that provide resistance to naturally occurring antibiotics in a competitive environmental niche, or the proteins produced may act as toxins
under similar circumstances, or allow the organism to utilize
particular organic compounds that would be advantageous when nutrients
are scarce.
History
The term plasmid was introduced in 1952 by the American molecular biologist Joshua Lederberg to refer to "any extrachromosomal hereditary determinant."
The term's early usage included any bacterial genetic material that
exists extrachromosomally for at least part of its replication cycle,
but because that description includes bacterial viruses, the notion of
plasmid was refined over time to comprise genetic elements that
reproduce autonomously.
Later in 1968, it was decided that the term plasmid should be adopted as the term for extrachromosomal genetic element,
and to distinguish it from viruses, the definition was narrowed to
genetic elements that exist exclusively or predominantly outside of the
chromosome and can replicate autonomously.
Properties and characteristics
There
are two types of plasmid integration into a host bacteria:
Non-integrating plasmids replicate as with the top instance, whereas episomes, the lower example, can integrate into the host chromosome.
In order for plasmids to replicate independently within a cell, they must possess a stretch of DNA that can act as an origin of replication. The self-replicating unit, in this case the plasmid, is called a replicon.
A typical bacterial replicon may consist of a number of elements, such
as the gene for plasmid-specific replication initiation protein (Rep),
repeating units called iterons, DnaA boxes, and an adjacent AT-rich region.
Smaller plasmids make use of the host replicative enzymes to make
copies of themselves, while larger plasmids may carry genes specific for
the replication of those plasmids. A few types of plasmids can also
insert into the host chromosome, and these integrative plasmids are
sometimes referred to as episomes in prokaryotes.
Plasmids almost always carry at least one gene. Many of the
genes carried by a plasmid are beneficial for the host cells, for
example: enabling the host cell to survive in an environment that would
otherwise be lethal or restrictive for growth. Some of these genes
encode traits for antibiotic resistance or resistance to heavy metal,
while others may produce virulence factors
that enable a bacterium to colonize a host and overcome its defences,
or have specific metabolic functions that allow the bacterium to utilize
a particular nutrient, including the ability to degrade recalcitrant or
toxic organic compounds. Plasmids can also provide bacteria with the ability to fix nitrogen.
Some plasmids, however, have no observable effect on the phenotype of
the host cell or its benefit to the host cells cannot be determined, and
these plasmids are called cryptic plasmids.
Naturally occurring plasmids vary greatly in their physical
properties. Their size can range from very small mini-plasmids of less
than a 1 kilobase pairs (Kbp), to very large megaplasmids of several
megabase pairs (Mbp). At the upper end, little can differentiate
between a megaplasmid and a minichromosome.
Plasmids are generally circular, but examples of linear plasmids are
also known. These linear plasmids require specialized mechanisms to
replicate their ends.
Plasmids may be present in an individual cell in varying number,
ranging from one to several hundreds. The normal number of copies of
plasmid that may be found in a single cell is called the copy number,
and is determined by how the replication initiation is regulated and
the size of the molecule. Larger plasmids tend to have lower copy
numbers. Low-copy-number plasmids that exist only as one or a few copies in each bacterium are, upon cell division,
in danger of being lost in one of the segregating bacteria. Such
single-copy plasmids have systems that attempt to actively distribute a
copy to both daughter cells. These systems, which include the parABS system and parMRC system, are often referred to as the partition system or partition function of a plasmid.
Classifications and types
Overview of bacterial conjugation
Electron micrograph of a DNA fiber bundle, presumably of a single bacterial chromosome loop.
Electron micrograph of a bacterial DNA plasmid (chromosome fragment).
Plasmids may be classified in a number of ways. Plasmids can be
broadly classified into conjugative plasmids and non-conjugative
plasmids. Conjugative plasmids contain a set of transfer or tra genes which promote sexual conjugation between different cells. In the complex process of conjugation, plasmid may be transferred from one bacterium to another via sex pili encoded by some of the tra genes (see figure).
Non-conjugative plasmids are incapable of initiating conjugation, hence
they can be transferred only with the assistance of conjugative
plasmids. An intermediate class of plasmids are mobilizable, and carry
only a subset of the genes required for transfer. They can parasitize a
conjugative plasmid, transferring at high frequency only in its
presence.
Plasmids can also be classified into incompatibility groups. A
microbe can harbour different types of plasmids, but different plasmids
can only exist in a single bacterial cell if they are compatible. If
two plasmids are not compatible, one or the other will be rapidly lost
from the cell. Different plasmids may therefore be assigned to different
incompatibility groups depending on whether they can coexist together.
Incompatible plasmids (belonging to the same incompatibility group)
normally share the same replication or partition mechanisms and can thus
not be kept together in a single cell.
Another way to classify plasmids is by function. There are five main classes:
- Fertility F-plasmids, which contain tra genes. They are capable of conjugation and result in the expression of sex pili.
- Resistance (R) plasmids, which contain genes that provide resistance against antibiotics or poisons. Historically known as R-factors, before the nature of plasmids was understood.
- Col plasmids, which contain genes that code for bacteriocins, proteins that can kill other bacteria.
- Degradative plasmids, which enable the digestion of unusual substances, e.g. toluene and salicylic acid.
- Virulence plasmids, which turn the bacterium into a pathogen.
Plasmids can belong to more than one of these functional groups.
Vectors
Artificially constructed plasmids may be used as vectors in genetic engineering.
These plasmids serve as important tools in genetics and biotechnology
labs, where they are commonly used to clone and amplify (make many
copies of) or express particular genes.
A wide variety of plasmids are commercially available for such uses.
The gene to be replicated is normally inserted into a plasmid that
typically contains a number of features for their use. These include a
gene that confers resistance to particular antibiotics (ampicillin is most frequently used for bacterial strains), an origin of replication to allow the bacterial cells to replicate the plasmid DNA, and a suitable site for cloning (referred to as a multiple cloning site).
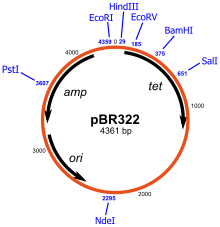
A schematic representation of the pBR322 plasmid, one of the first plasmids to be used widely as a cloning vector. Shown on the plasmid diagram are the genes encoded (amp and tet for ampicillin and tetracycline resistance respectively), its origin of replication (ori), and various restriction sites (indicated in blue).
Cloning
Plasmids are the most-commonly used bacterial cloning vectors. These cloning vectors contain a site that allows DNA fragments to be inserted, for example a multiple cloning site or polylinker which has several commonly used restriction sites to which DNA fragments may be ligated. After the gene of interest is inserted, the plasmids are introduced into bacteria by a process called transformation. These plasmids contain a selectable marker,
usually an antibiotic resistance gene, which confers on the bacteria an
ability to survive and proliferate in a selective growth medium
containing the particular antibiotics. The cells after transformation
are exposed to the selective media, and only cells containing the
plasmid may survive. In this way, the antibiotics act as a filter to
select only the bacteria containing the plasmid DNA. The vector may
also contain other marker genes or reporter genes
to facilitate selection of plasmid with cloned insert. Bacteria
containing the plasmid can then be grown in large amounts, harvested,
and the plasmid of interest may then be isolated using various methods
of plasmid preparation.
A plasmid cloning vector is typically used to clone DNA fragments of up to 15 kbp. To clone longer lengths of DNA, lambda phage with lysogeny genes deleted, cosmids, bacterial artificial chromosomes, or yeast artificial chromosomes are used.
Protein production
Another major use of plasmids is to make large amounts of proteins.
In this case, researchers grow bacteria containing a plasmid harboring
the gene of interest. Just as the bacterium produces proteins to confer
its antibiotic resistance, it can also be induced to produce large
amounts of proteins from the inserted gene. This is a cheap and easy
way of mass-producing the protein the gene codes for, for example, insulin.
Gene therapy
Plasmid may also be used for gene transfer into human cells as potential treatment in gene therapy so that it may express the protein that is lacking in the cells. Some strategies of gene therapy require the insertion of therapeutic genes at pre-selected chromosomal target sites within the human genome. Plasmid vectors are one of many approaches that could be used for this purpose. Zinc finger nucleases (ZFNs) offer a way to cause a site-specific double-strand break to the DNA genome and cause homologous recombination.
Plasmids encoding ZFN could help deliver a therapeutic gene to a
specific site so that cell damage, cancer-causing mutations, or an
immune response is avoided.
Disease models
Plasmids
were historically used to genetically engineer the embryonic stem cells
of rats in order to create rat genetic disease models. The limited
efficiency of plasmid-based techniques precluded their use in the
creation of more accurate human cell models. However, developments in Adeno-associated virus recombination techniques, and Zinc finger nucleases, have enabled the creation of a new generation of isogenic human disease models.
Episomes
The term episome was introduced by François Jacob and Élie Wollman
in 1958 to refer to extra-chromosomal genetic material that may
replicate autonomously or become integrated into the chromosome. Since the term was introduced, however, its use has shifted, as plasmid
has become the preferred term for autonomously replicating
extrachromosomal DNA. At a 1968 symposium in London some participants
suggested that the term episome be abandoned, although others continued to use the term with a shift in meaning.
Today some authors use episome in the context of
prokaryotes to refer to a plasmid that is capable of integrating into
the chromosome. The integrative plasmids may be replicated and stably
maintained in a cell through multiple generations, but always at some
stage they exist as an independent plasmid molecule. In the context of eukaryotes, the term episomes is used to mean a non-integrated extrachromosomal closed circular DNA molecule that may be replicated in the nucleus. Viruses are the most common examples of this, such as herpesviruses, adenoviruses, and polyomaviruses, but some are plasmids. Other examples include aberrant chromosomal fragments, such as double minute chromosomes,
that can arise during artificial gene amplifications or in pathologic
processes (e.g., cancer cell transformation). Episomes in eukaryotes
behave similarly to plasmids in prokaryotes in that the DNA is stably
maintained and replicated with the host cell. Cytoplasmic viral
episomes (as in poxvirus infections) can also occur. Some episomes, such as herpesviruses, replicate in a rolling circle mechanism, similar to bacterial phage viruses. Others replicate through a bidirectional replication mechanism (Theta type plasmids). In either case, episomes remain physically separate from host cell chromosomes. Several cancer viruses, including Epstein-Barr virus and Kaposi's sarcoma-associated herpesvirus, are maintained as latent, chromosomally distinct episomes in cancer cells, where the viruses express oncogenes
that promote cancer cell proliferation. In cancers, these episomes
passively replicate together with host chromosomes when the cell
divides. When these viral episomes initiate lytic replication to generate multiple virus particles, they in general activate cellular innate immunity defense mechanisms that kill the host cell.
Plasmid maintenance
Some plasmids or microbial hosts include an addiction system or postsegregational killing system (PSK), such as the hok/sok (host killing/suppressor of killing) system of plasmid R1 in Escherichia coli. This variant produces both a long-lived poison and a short-lived antidote. Several types of plasmid addiction systems (toxin/ antitoxin, metabolism-based, ORT systems) were described in the literature
and used in biotechnical (fermentation) or biomedical (vaccine therapy)
applications. Daughter cells that retain a copy of the plasmid survive,
while a daughter cell that fails to inherit the plasmid dies or suffers
a reduced growth-rate because of the lingering poison from the parent
cell. Finally, the overall productivity could be enhanced.
In contrast, virtually all biotechnologically used plasmids (such
as pUC18, pBR322 and derived vectors) do not contain toxin-antitoxin
addiction systems and thus need to be kept under antibiotic pressure to
avoid plasmid loss.
Yeast plasmids
Yeasts naturally harbour various plasmids. Notable among them are 2 µm plasmids — small circular plasmids often used for genetic engineering of yeast — and linear pGKL plasmids from Kluyveromyces lactis, that are responsible for killer phenotypes.
Other types of plasmids are often related to yeast cloning vectors that include:
- Yeast integrative plasmid (YIp), yeast vectors that rely on integration into the host chromosome for survival and replication, and are usually used when studying the functionality of a solo gene or when the gene is toxic. Also connected with the gene URA3, that codes an enzyme related to the biosynthesis of pyrimidine nucleotides (T, C);
- Yeast Replicative Plasmid (YRp), which transport a sequence of chromosomal DNA that includes an origin of replication. These plasmids are less stable, as they can get lost during the budding.
Plasmid DNA extraction
As
alluded to above, plasmids are often used to purify a specific
sequence, since they can easily be purified away from the rest of the
genome. For their use as vectors, and for molecular cloning, plasmids often need to be isolated.
There are several methods to isolate plasmid DNA from bacteria, the archetypes of which are the miniprep and the maxiprep/bulkprep.
The former can be used to quickly find out whether the plasmid is
correct in any of several bacterial clones. The yield is a small amount
of impure plasmid DNA, which is sufficient for analysis by restriction digest and for some cloning techniques.
In the latter, much larger volumes of bacterial suspension are
grown from which a maxi-prep can be performed. In essence, this is a
scaled-up miniprep followed by additional purification. This results in
relatively large amounts (several hundreds micrograms) of very pure
plasmid DNA.
In recent times, many commercial kits have been created to
perform plasmid extraction at various scales, purity, and levels of
automation. Commercial services can prepare plasmid DNA at quoted prices
below $300/mg in milligram quantities and $15/mg in gram quantities
(early 2007).
Conformations
Plasmid DNA may appear in one of five conformations, which (for a given size) run at different speeds in a gel during electrophoresis.
The conformations are listed below in order of electrophoretic mobility
(speed for a given applied voltage) from slowest to fastest:
- Nicked open-circular DNA has one strand cut.
- Relaxed circular DNA is fully intact with both strands uncut, but has been enzymatically relaxed (supercoils removed). This can be modeled by letting a twisted extension cord unwind and relax and then plugging it into itself.
- Linear DNA has free ends, either because both strands have been cut or because the DNA was linear in vivo. This can be modeled with an electrical extension cord that is not plugged into itself.
- Supercoiled (or covalently closed-circular) DNA is fully intact with both strands uncut, and with an integral twist, resulting in a compact form. This can be modeled by twisting an extension cord and then plugging it into itself.
- Supercoiled denatured DNA is like supercoiled DNA, but has unpaired regions that make it slightly less compact; this can result from excessive alkalinity during plasmid preparation.
The rate of migration for small linear fragments is directly
proportional to the voltage applied at low voltages. At higher voltages,
larger fragments migrate at continuously increasing yet different
rates. Thus, the resolution of a gel decreases with increased voltage.
At a specified, low voltage, the migration rate of small linear
DNA fragments is a function of their length. Large linear fragments
(over 20 kb or so) migrate at a certain fixed rate regardless of length.
This is because the molecules 'resperate', with the bulk of the
molecule following the leading end through the gel matrix. Restriction digests
are frequently used to analyse purified plasmids. These enzymes
specifically break the DNA at certain short sequences. The resulting
linear fragments form 'bands' after gel electrophoresis.
It is possible to purify certain fragments by cutting the bands out of
the gel and dissolving the gel to release the DNA fragments.
Because of its tight conformation, supercoiled DNA migrates faster through a gel than linear or open-circular DNA.
Software for bioinformatics and design
The use of plasmids as a technique in molecular biology is supported by bioinformatics software. These programs record the DNA sequence of plasmid vectors, help to predict cut sites of restriction enzymes, and to plan manipulations. Examples of software packages that handle plasmid maps are ApE, Clone Manager, GeneConstructionKit, Geneious, Genome Compiler, LabGenius, Lasergene, MacVector, pDraw32, Serial Cloner, VectorFriends, Vector NTI, and WebDSV. These software help conduct entire experiments in silico before doing wet experiments.
Plasmid collections
Many
plasmids have been created over the years and researchers have given
out plasmids to plasmid databases such as the non-profit organisations Addgene and BCCM/LMBP. One can find and request plasmids from those databases in order to continue research.
Researcher also often upload sequences of plasmids in the NCBI database. Using the NCBI database, sequences of specific plasmids can be looked up.