A series of biochemical switches control transitions between and within the various phases of the cell cycle.
The cell cycle is a series of complex, ordered, sequential events that
control how a single cell divides into two cells, and involves several
different phases. The phases include the G1 and G2 phases, DNA replication or S phase, and the actual process of cell division, mitosis or M phase. During the M phase, the chromosomes separate and cytokinesis occurs.
The switches maintain the orderly progression of the cell cycle
and act as checkpoints to ensure that each phase has been properly
completed before progression to the next phase. For example, Cdk, or cyclin dependent kinase,
is a major control switch for the cell cycle and it allows the cell to
move from G1 to S or G2 to M by adding phosphate to protein substrates.
Such multi-component (involving multiple inter-linked proteins) switches
have been shown to generate decisive, robust (and potentially
irreversible) transitions and trigger stable oscillations.
As a result, they are a subject of active research that tries to
understand how such complex properties are wired into biological control
systems.
Feedback loops
Many biological circuits produce complex outputs by exploiting one or more feedback
loops. In a sequence of biochemical events, feedback would refer to a
downstream element in the sequence (B in the adjacent image) affecting
some upstream component (A in the adjacent image) to affect its own
production or activation (output) in the future. If this element acts to
enhance its own output, then it engages in positive feedback
(blue arrow). A positive feedback loop is also known as a
self-reinforcing loop, and it is possible that these loops can be part
of a larger loop, as this is characteristic of regulatory circuits.
Conversely, if this element leads to its own inhibition through upstream elements, this is canonically negative feedback
(red blunt arrow). A negative feedback loop is also known as a
balancing loop, and it may be common to see oscillations in which a
delayed negative feedback signal is used to maintain homeostatic balance
in the system.
Feedback loops can be used for amplification (positive) or
self-correction (negative). The right combination of positive and
negative feedback loops can generate ultrasensitivity and bistability, which in turn can generate decisive transitions and oscillations.
Combination of positive and negative feedback loops
Positive
and negative feedback loops do not always operate distinctly. In the
mechanism of biochemical switches, they work together to create a
flexible system. For example, according to Pfeuty & Kaneko (2009),
to overcome a drawback in biochemical systems, positive feedback
regulation loops may interact with negative regulation loops to
facilitate escape from stable states. The coexistence of two stable states is known as bistability, which is often the result of positive feedback regulations.
An example that reveals the interaction of the multiple negative
and positive feedback loops is the activation of cyclin-dependent
protein kinases, or Cdks14. Positive feedback loops play a role by
switching cells from low to high Cdk-activity. The interaction between
the two types of loops is evident in mitosis. While positive feedback
initiates mitosis, a negative feedback loop promotes the inactivation of
the cyclin-dependent kinases by the anaphase-promoting complex. This
example clearly shows the combined effects that positive and negative
feedback loops have on cell-cycle regulation.
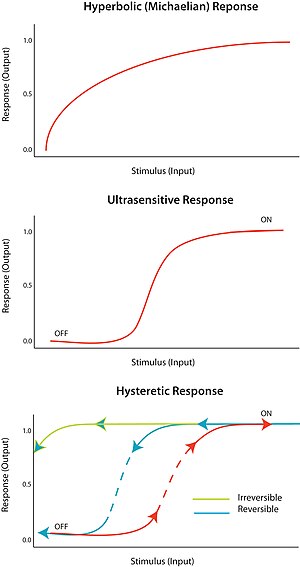
Ultrasensitivity
An "all-or-none" response to a stimulus is termed ultrasensitivity.
In other words, a very small change in stimulus causes a very large
change in response, producing a sigmoidal dose-response curve. An
ultrasensitive response is described by the general equation V = Sn/(Sn + Km), known as the Hill equation,
when n, the Hill coefficient, is more than 1. The steepness of the
sigmoidal curve depends on the value of n. A value of n = 1 produces a
hyperbolic or Michaelian response. Ultrasensitivity is achieved in a
variety of systems; a notable example is the cooperative binding of the
enzyme hemoglobin
to its substrate. Since an ultrasensitive response is almost ‘digital’,
it can be used to amplify a response to a stimulus or cause a decisive
sharp transition (between ‘off’ and ‘on’ states).
Ultrasensitivity plays a large role in cell-cycle regulation. For
example, Cdk1 and Wee1 are mitotic regulators, and they are able to
inactivate each other through inhibitory phosphorylation. This
represents a double negative feedback loop in which both regulators
inactivate each other. According to Kim et al. (2007), there must be an
ultrasensitive element to generate a bistable response. It turns out
that Wee1 has an ultrasensitive response to Cdk1, and this likely arises
because of substrate competition among the various phosphorylation
sites on Wee1.
Bistability
Bistability
implies hysteresis, and hysteresis implies multistability.
Multistability indicates the presence of two or more stable states for a
given input. Therefore, bistability is the ability of a system to exist in two steady states.
In other words, there is a range of stimulus values for which the
response can have two steady-state values. Bistability is accompanied by
hysteresis,
which means that the system approaches one of the two steady states
preferentially depending on its history. Bistability requires feedback
as well as an ultrasensitive circuit element.
Under the proper circumstances, positive and negative feedback
loops can provide the conditions for bistability; for example, by having
positive feedback coupled to an ultrasensitive response element with
the circuit. A hysteretic bistable system can act as a robust reversible
switch because it is harder for the system to transition between ‘on’
and ‘off’ states (compared to the equivalent monostable ultransensitive
response). The system could also be poised such that one of the
transitions is physically unattainable; for example, no amount of
reduction in the stimulus will return the system to the ‘off’-state once
it is already in the ‘on’ state. This would form a robust irreversible
switch.
There is no one-to-one correspondence between network topology,
since many networks have a similar input and output relationship. A
network topology does not imply input or output, and similarly input or
output does not imply network topology. It is for this reason that
parameterization is very important for circuit function. If the dynamics
of the input are comparable or faster than the response of the system,
the response may appear hysteretic.
Three cell cycle switches are described below that achieve abrupt
and/or irreversible transitions by exploiting some of the mechanisms
described above.
The G1/S switch
The G1/S transition,
more commonly known as the Start checkpoint in budding yeast (the
restriction point in other organisms) regulates cell cycle commitment.
At this checkpoint, cells either arrest before DNA replication (due to
limiting nutrients or a pheromone signal), prolong G1 (size control), or
begin replication and progress through the rest of the cell cycle. The
G1/S regulatory network or regulon in budding yeast includes the G1
cyclins Cln1, Cln2 and Cln3, Cdc28 (Cdk1), the transcription factors SBF
and MBF, and the transcriptional inhibitor Whi5.
Cln3 interacts with Cdk1 to initiate the sequence of events by
phosphorylating a large number of targets, including SBF, MBF and Whi5.
Phosphorylation of Whi5 causes it to translocate out of the nucleus,
preventing it from inhibiting SBF and MBF. Active SBF/MBF drive the G1/S
transition by turning on the B-type cyclins and initiating DNA
replication, bud formation and spindle body duplication. Moreover,
SBF/MBF drives expression of Cln1 and Cln2, which can also interact with
Cdk1 to promote phosphorylation of its targets.
This G1/S switch was initially thought to function as a linear sequence of events starting with Cln3 and ending in S phase.
However, the observation that any one of the Clns was sufficient to
activate the regulon indicated that Cln1 and Cln2 might be able to
engage positive feedback to activate their own transcription. This would
result in a continuously accelerating cycle that could act as an
irreversible bistable trigger. Skotheim et al. used single-cell measurements in budding yeast to show that this positive feedback does indeed occur.
A small amount of Cln3 induces Cln1/2 expression and then the feedback
loop takes over, leading to rapid and abrupt exit of Whi5 from the
nucleus and consequently coherent expression of G1/S regulon genes. In
the absence of coherent gene expression, cells take longer to exit G1
and a significant fraction even arrest before S phase, highlighting the
importance of positive feedback in sharpening the G1/S switch.
The G1/S cell cycle checkpoint controls the passage of eukaryotic
cells from the first gap phase, G1, into the DNA synthesis phase, S. In
this switch in mammalian cells, there are two cell cycle kinases that
help to control the checkpoint: cell cycle kinases CDK4/6-cyclin D and
CDK2-cyclin E.
The transcription complex that includes Rb and E2F is important in
controlling this checkpoint. In the first gap phase, the Rb-HDAC
repressor complex binds to the E2F-DP1 transcription factors, therefore
inhibiting the downstream transcription. The phosphorylation of Rb by
CDK4/6 and CDK2 dissociates the Rb-repressor complex and serves as an
on/off switch for the cell cycle. Once Rb is phosphorylated, the
inhibition is released on the E2F transcriptional activity. This allows
for the transcription of S phase genes encoding for proteins that
amplify the G1 to S phase switch.
Many different stimuli apply checkpoint controls including TGFb,
DNA damage, contact inhibition, replicative senescence, and growth
factor withdrawal. The first four act by inducing members of the INK4 or
Kip/Cip families of cell cycle kinase inhibitors. TGFb inhibits the
transcription of Cdc25A, a phosphatase that activates the cell cycle
kinases, and growth factor withdrawal activates GSK3b, which
phosphorylates cyclin D. This leads to its rapid ubiquitination.
The G2/M switch
G2
is commenced by E2F-mediated transcription of cyclin A, which forms the
cyclin A-Cdk2 complex. In order to proceed into mitosis, the cyclin B-Cdk1
complex (first discovered as MPF or M-phase promoting factor; Cdk1 is
also known as Cdc2 in fission yeast and Cdc28 in budding yeast) is
activated by Cdc25, a protein phosphatase.
As mitosis starts, the nuclear envelope disintegrates, chromosomes
condense and become visible, and the cell prepares for division. Cyclin
B-Cdk1 activation results in nuclear envelope breakdown, which is a
characteristic of the initiation of mitosis.
The cyclin B-Cdk1 complex participates in a regulatory circuit in
which Cdk1 can phosphorylate and activate its activator, Cdc25
(positive feedback), and phosphorylate and inactivate its inactivator,
the kinase Wee1 (double-negative feedback). This circuit could act as a bistable trigger
with one stable steady state in G2 (Cdk1 and Cdc25 off, Wee1 on) and a
second stable steady state in M phase (Cdk1 and Cdc25 active, Wee1 off).
However, Wee1 is itself regulated by other factors, such as Cdr2.
It was suggested and defended by Jin et al.
in their series of experiments with the human HeLa cell line in 1998
that it is the spatial location of cyclin B within the cell that
initiates mitosis. Known from previous experiments in both human cells
and starfish oocytes, Jin et al. summarize that cyclin B1 is abundant in
the cytoplasm during non-dividing phases of mitosis, but is identified
in the nucleus, in complex with Cdk1, immediately before the cell enters
mitosis. Other experimenters showed that cells would not divide if
cyclin B remains in the cytoplasm. In order to further investigate the
effect of spatial location of cyclin B on cell division and cycle
control, Jin et al. tagged cyclin B with a nuclear localization signal
(NLS) that would keep the cyclin within the nucleus. Initially, this NLS
cyclin B did not induce the expected effect of accelerated mitotic
entry. This result is due to the inhibition detailed in the figure
below. Wee1, an inhibitor on the cyclin B-Cdk1 complex, is localized in
the nucleus, and likely phosphorylating the NLS cyclin B, rendering it
unable to perform as predicted. This postulation was confirmed when Jin
et al. employed Cdc2AF, an unphosphorylatable mutant of Cdk1, and saw
accelerated entry to cell division due to the nuclear localization of
the cyclin B. Therefore, nuclear localization of cyclin B is necessary
but not sufficient to trigger cell division.
In investigation of cell cycle regulation, Jin et al. manipulated
cells in order to evaluate the localization of cyclin B in cells with
DNA damage. Through combination of DNA damage and nuclear localization
of exogenous cyclin B, they were able to determine that cells would
divide even with DNA damage if the cyclin B were forced to be expressed
in the nucleus. This suggests that spatial localization of cyclin B may
play a role as a checkpoint of mitosis. If the cells, under normal
circumstances, don’t divide when their genetic information is damaged,
but will enter mitosis if endogenous cyclin B is expressed in the
nucleus, it is likely that the translocation of the cyclin B to the
cytoplasm is a mechanism that prevents immature mitotic entry. This
hypothesis was further supported by Jin et al.’s analysis of cells
arrested in G2 due to DNA damage. In these cells, Jin et al. observed
high levels of cyclin B-Cdc2 complex activity in the cytoplasm. This is
supporting evidence for the previously mentioned theory because it shows
that the Cdc2 can activate the cyclin without immediate translocation
to the nucleus. Additionally, the accumulation of cyclin B-Cdk1
complexes in the cytoplasm of cells that are not dividing due to DNA
damage supports the theory that it is nuclear localization of cyclin B
that initiates mitotic entry.
To conclude, spatial localization of cyclin B plays a role in
mitotic entry. Translocation of cyclin B from the cytoplasm to the
nucleus is necessary for cell division, but not sufficient, as its
inhibitors do not allow the cell to enter mitosis prematurely. In
addition to the back up inhibition of the cyclin B-Cdk1 complex,
premature cellular division is prevented by the translocation of the
cyclin B itself. The cyclin B-Cdk1 complex will remain in the cytoplasm
in cells with DNA damage, rather than translocate to the nucleus,
keeping the cell inhibiting the cell from entering mitosis. The next
question addressed by researchers in this field is by which specific
mechanism is this translocation regulated.
Santos et al.
hypothesized that the translocation of cyclin B is regulated by a
mechanism of positive feedback, similar to that which regulates the
activation of the cyclin B-Cdk1 complex. They believed that the positive
feedback loop involves the phosphorylation of the cyclin B and its
translocation to the nucleus. To begin to investigate this, they first
reconfirmed some of the results of the Jin et al. experiments, utilizing
immunofluorescence to show cyclin B in the cytoplasm prior to division,
and translocation to the nucleus to initiate mitosis, which they
operationalized by comparing relative to nuclear envelope breakdown
(NEB). Using nuclear cyclin that cannot be inactivated by Wee1 or Myt1,
Santos et al. observed that active nuclear cyclin recruits more cyclin
from the cytoplasm to be translocated to the nucleus. They confirmed
this observation by employing a rapamycin treatment, iRap. iRap induces
translocation of tagged cyclin B from the cytoplasm to the nucleus.
Remarkably, Santos et al. saw that untagged cyclin B migrated with the
cyclin B influenced by iRap. The untagged cyclin is insentive to the
treatment, and moves independently from the treated cyclin. This
supports the first part of the positive feedback loop, that nuclear
localization of cyclin B, which leads to mitotic entry, promotes
increased translocation of cytoplasmic cyclin B to the nucleus, further
promoting the remaining cytoplasmic cyclin B to migrate to the nucleus,
etc.
Santos et al. further hypothesize that phosphorylation of the
cyclin B is another component of the positive feedback loop. They
observed that the cyclin B naturally enters the nucleus before NEB. In
contrast, mutated, unphosphorylatable cyclin B enters the nucleus during
NEB. This is unexpected because it is characteristic of the cell cycle
for the cyclin to translocate to the nucleus prior to NEB in order to
induce cell cycle progression into mitotic division. Therefore, Santos
et al. conclude that the phosphorylation of the cyclin B promotes the
translocation to the nucleus. However, in addition, translocation to the
nucleus promotes phosphorylation of the cyclin. It is noted by the
authors that phosphorylation of cyclin B is nineteen times more
favorable in the nucleus than in the cytoplasm, due to the smaller
overall volume of the nucleus, allowing a faster phosphorylation rate.
The increased translocation due to phosphorylation and increased
phosphorylation due to translocation exemplify the positive feedback
loop that resembles that previously discovered, which activates the
cyclin B-Cdk1 complex.
In conclusion, nuclear localization of cyclin B is necessary for
cellular entry into mitosis. The translocation of the cyclin from the
cytoplasm to the nucleus, which allows for cellular division, is
regulated by a positive feedback loop. Active cyclin B translocates to
the nucleus and promotes activation and translocation of additional
units of cyclin residing in the nucleus. This phenomenon is enhanced
when considering phosphorylation. Phosphorylation of cyclin B promotes
translocation to the nucleus, and cyclin B in the nucleus is much more
likely to be phosphorylated, so nuclear localization promotes cyclin B
phosphorylation in return.
Once cells are in mitosis, cyclin B-Cdk1 activates the anaphase-promoting complex
(APC), which in turn inactivates cyclin B-Cdk1 by degrading cyclin B,
eventually leading to exit from mitosis. Coupling the bistable Cdk1
response function to the negative feedback from the APC could generate
what is known as a relaxation oscillator,
with sharp spikes of Cdk1 activity triggering robust mitotic cycles.
However, in a relaxation oscillator, the control parameter moves slowly
relative to the system’s response dynamics which may be an accurate
representation of mitotic entry, but not necessarily mitotic exit.
It is necessary to inactivate the cyclin B-Cdk1 complex in order
to exit the mitotic stage of the cell cycle. The cells can then return
to the first gap phase G1 and wait until the cycle proceeds yet again.
In 2003 Pomerening et al. provided strong evidence for this
hypothesis by demonstrating hysteresis and bistability in the activation
of Cdk1 in the cytoplasmic extracts of Xenopus oocytes.
They first demonstrated a discontinuous sharp response of Cdk1 to
changing concentrations of non-destructible Cyclin B (to decouple the
Cdk1 response network from APC-mediated negative feedback). However,
such a response would be consistent with both a monostable,
ultrasensitive transition and a bistable transition. To distinguish
between these two possibilities, they measured the steady-state levels
of active Cdk1 in response to changing cyclin levels, but in two
separate experiments, one starting with an interphase
extract and one starting with an extract already in mitosis. At
intermediate concentrations of cyclin they found two steady-state
concentrations of active Cdk1. Which of the two steady states was
occupied depended on the history of the system, i.e.whether they started
with interphase or mitotic extract, effectively demonstrating
hysteresis and bistability.
In the same year, Sha et al.
independently reached the same conclusion revealing the hysteretic loop
also using Xenopus laevis egg extracts. In this article, three
predictions of the Novak-Tyson model were tested in an effort to
conclude that hysteresis is the driving force for "cell-cycle
transitions into and out of mitosis". The predictions of the Novak-Tyson
model are generic to all saddle-node bifurcations. Saddle-node
bifurcations are extremely useful bifurcations in an imperfect world
because they help describe biological systems which are not perfect. The
first prediction was that the threshold concentration of cyclin to
enter mitosis is higher than the threshold concentration of cyclin to
exit mitosis, and this was confirmed by supplementing cycling egg
extracts with non-degradable cyclin B and measuring the activation and
inactivation threshold after the addition of cycloheximide (CHX), which
is a protein synthesis inhibitor.
Furthermore, the second prediction of the Novak-Tyson model was also
validated: unreplicated deoxyribonucleic acid, or DNA, increases the
threshold concentration of cyclin that is required to enter mitosis. In
order to arrive at this conclusion, cytostatic factor released extracts
were supplemented with CHX, APH (a DNA polymerase inhibitor), or both,
and non-degradable cyclin B was added. The third and last prediction
that was tested and proven true in this article was that the rate of
Cdc2 activation slows down near the activation threshold concentration
of cyclin. These predictions and experiments demonstrate the toggle-like
switching behavior that can be described by hysteresis in a dynamical
system.
Metaphase-anaphase switch
In the transition from metaphase to anaphase, it is crucial that sister chromatids are properly and simultaneously separated to opposite ends of the cell.
Separation of sister-chromatids is initially strongly inhibited to
prevent premature separation in late mitosis, but this inhibition is
relieved through destruction of the inhibitory elements by the anaphase-promoting complex (APC) once sister-chromatid bi-orientation is achieved. One of these inhibitory elements is securin, which prevents the destruction of cohesin, the complex that holds the sister-chromatids together, by binding the protease separase which targets Scc1, a subunit of the cohesin complex, for destruction. In this system, the phosphatase Cdc14
can remove an inhibitory phosphate from securin, thereby facilitating
the destruction of securin by the APC, releasing separase. As shown by
Uhlmann et al., during the attachment of chromosomes to the mitotic
spindle the chromatids remain paired because cohesion between the
sisters prevents separation.
Cohesion is established during DNA replication and depends on cohesin,
which is a multisubunit complex composed of Scc1, Scc3, Smc2, and Smc3.
In yeast at the metaphase-to-anaphase transition, Scc1 dissociates from
the chromosomes and the sister chromatids separate. This action is
controlled by the Esp1 protein, which is tightly bound by the anaphase
inhibitor Pds1 that is destroyed by the anaphase-promoting complex. In
order to verify that Esp1 does play a role in regulating Scc1 chromosome
association, cell strains were arrested in G1 with an alpha factor.
These cells stayed in arrest during the development. Esp1-1 mutant cells
were used and the experiment was repeated, and Scc1 successfully bound
to the chromosomes and remained associated even after the synthesis was
terminated. This was crucial in showing that with Esp1, Scc1 is hindered
in its ability to become stably associated with chromosomes during G1,
and Esp1 can in fact directly remove Scc1 from chromosomes.
It has been shown by Holt et al.
that separase activates Cdc14, which in turn acts on securin, thus
creating a positive feedback loop that increases the sharpness of the
metaphase to anaphase transition and coordination of sister-chromatid
separation.
Holt et al. probed the basis for the effect of positive feedback in
securin phosphorylation by using mutant 'securin' strains of yeast, and
testing how changes in the phosphoregulation of securin affects the
synchrony of sister chromatid separation. Their results indicate that
interfering with this positive securin-separase-cdc14 loop decreases
sister chromatid separation synchrony. This positive feedback can
hypothetically generate bistability in the transition to anaphase,
causing the cell to make the irreversible decision to separate
sister-chromatids.
Mitotic exit
Mitotic exit is an important transition point that signifies the end of mitosis and the onset of new G1 phase
for a cell, and the cell needs to rely on specific control mechanisms
to ensure that once it exits mitosis, it never returns to mitosis until
it has gone through G1, S, and G2 phases and passed all the necessary
checkpoints. Many factors including cyclins, cyclin-dependent kinases (CDKs), ubiquitin ligases, inhibitors of cyclin-dependent kinases, and reversible phosphorylations regulate mitotic exit to ensure that cell cycle events occur in correct order with the fewest errors. The end of mitosis is characterized by spindle breakdown, shortened kinetochore microtubules, and pronounced outgrowth of astral (non-kinetochore) microtubules. For a normal eukaryotic cell, mitotic exit is irreversible.
Proteolytic degradation
Fig. 1
Immunofluorescence patterns of cyclin B and phosphorylated
cyclin-dependent kinase1 (Cdk1) in HeLa cells change as they go from G2
to anaphase.
Many speculations were made with regard to the control mechanisms
employed by a cell to promote the irreversibility of mitotic exit in a
eukaryotic model organism, the budding yeast Saccharomyces cerevisiae.
Proteolytic degradation of cell cycle regulators and corresponding
effects on the levels of cyclin-dependent kinases were proposed as a
mechanism that promotes eukaryotic cell cycle and metaphase-to-anaphase
transition in particular.
In this theory, anaphase promoting complex
(APC), a class of ubiquitin ligase, facilitates degradation of mitotic
cyclins (Clb2) and anaphase-inhibiting factors (PDS1, CUT2) to promote
mitotic exit.
APC ubiquitinates nine-amino acid motif known as the destruction box (D
box) in the NH2-terminal domain of mitotic cyclins for degradation by
proteasome. APC in association with Cdc20
(APC-Cdc20) ubiquitinates and targets mitotic cyclins (Clb2) for
degradation at initial phase. Simultaneously, APC-Cdc20 mediates
degradation of securins, which inhibit separases
through binding, at anaphase onset. Released and active separase
cleaves cohesin that held sister chromatids together, facilitating
separation of sister chromatids and initiates mitotic exit by promoting
release of Cdc14 from nucleolus.
At later phase, downregulation of Cdk1 and activation of Cdc14, a
Cdh1-activating phosphatase, promotes formation of APC in association
with Cdh1 (APC-Cdh1) to degrade Clb2s. Cdc20 and Cdh1, which are the activators of APC, recruit substrates such as securin and B-type cyclins(Clb) for ubiquitination. Without Cdk1-Clb2 complexes to phosphorylate proteins that are involved in spindle dynamics such as Sli15, Ase1, and Ask1, spindle elongation and chromosomal segregation are promoted, facilitating mitotic exit.
The importance of proteolytic degradation in eukaryotic cell cycle
changed the view of cell division as a simple kinase cascade to a more
complex process in which interactions among phosphorylation,
ubiquitination, and proteolysis are necessary.
However, experiments using budding yeast cells with cdc28-as1, a
INM-PP1 (ATP analog)-sensitive Cdk allele, proved that destruction of
B-type cyclins (Clb) is not necessary for triggering irreversible
mitotic exit.
Clb2 degradation did shorten the Cdk1-inhibition period required for
triggering irreversible mitotic exit indicating that cyclin proteolysis
contributes to the dynamic nature of the eukaryotic cell cycle due to
slower timescale of its action but is unlikely to be the major
determining factor in triggering irreversible cell cycle transitions.
Sic1 levels
Discoveries
were made which indicated the importance of the level of the inhibitors
of cyclin-dependent kinases in regulating eukaryotic cell cycle. In
particular, the level of Sic1,
a stoichiometric inhibitor of Clb-CDK complexes in budding yeast, was
shown to be particularly important in irreversible G1-S transition by
irreversibly activating S phase kinases.
Sic1 level was shown to play a major role in triggering irreversible
mitotic exit (M-G1 transition) as well as in G1-S transition. During
mitosis, decreasing levels of Cdk1 leads to the activation of Cdc14, a
phosphatase that counteracts Cdk1 via activation of Cdh1 and Swi5, a
transcriptional activator of Sic1 proteins.
While degradation of Sic1 to a certain low level triggered the onset of
S phase, accumulation of Sic1 to a certain high level was required to
trigger irreversible mitotic exit.
Cdk1-inhibitors could induce mitotic exit even when degradation of
B-type cyclins was blocked by expression of non-degradable Clbs or
proteasome inhibitors. However, sister chromatids failed to segregate,
and cells reverted to mitosis once the inhibitors were washed away,
indicating that a threshold level of the inhibitors needs to be achieved
to trigger irreversible mitotic exit independently of cyclin
degradations.
Despite different thresholds of Sic1 level that are required to trigger
mitotic exit compared to G1-S transition, the level of Sic1 was shown
to play a key role in regulating eukaryotic cell cycle by inhibiting the
activity of CDKs.
Dynamical systems approach
Fig. 2
Irreversible and bistable switch in mitotic exit with control parameter
being Sic1 level and order parameter being cell cycle phases.
Because eukaryotic cell cycle involves a variety of proteins and
regulatory interactions, dynamical systems approach can be taken to
simplify a complex biological circuit into a general framework for
better analysis.
Among the four possible input/output relationships, the relationship
between Sic1 level and mitotic exit seems to show the characteristics of
an irreversible bistable switch, driven by feedback between APC-Cdh1,
Sic1, and Clb2-Cdk1. Bistability
is known to control biological functions such as cell cycle control and
cellular differentiation and play a key role in many cellular
regulatory networks.
Bistable input/output relationship is characterized by two stable
states with two bifurcation points. Multiple outputs are possible for
one specific input in the region of bistability, marked by two
bifurcation points. In addition, the bistable relationship displays
hysteresis: the final state/output depends on the history of the input
as well as the current value of input because the system has a memory.
One bifurcation point has a negative control parameter value (the
bifurcation point is on the other side of the axis), resulting in
disconnection between the two stable states and irreversibility of the
transition from one state to the other. With regard to mitotic exit, the
two stable states are defined by mitosis and G1 phase. Once Sic1 level
(input) accumulates beyond the threshold, irreversible transition occurs
from mitosis (stable state I) to G1 phase (stable state II).
In the imperfect environment, the only bifurcation that remains intact
is saddle-node bifurcation. Saddle-node bifurcation does not break down (saddle-node is the
expected generic behavior), while transcritical and pitchfork
bifurcations break down in the presence of imperfections. Thus, the only one-dimensional bifurcation that can exist in imperfect biological world is the saddle-node bifurcation.
The bistable relation between M-G1 transition and Sic1 level can be
represented as a diagram of two saddle-node bifurcations in which the
system’s behavior changes qualitatively with a small change in control
parameter, the amount of Sic1.
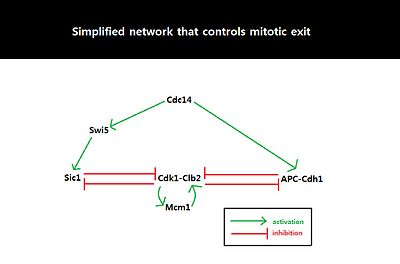
Systems-level feedback
Fig. 3
Simplified network involving Cdk1-Clb2, APC-Cdh1, Sic1, and Cdc14.
Double negative feedback loop, mediated by APC-Cdh1 and Sic1, is
required to suppress Cdk1-Clb2 and trigger mitotic exit.
Because the behavior of cell cycle critically depends on the amount of
Sic1 at the M-G1 transition state, the amount of Sic1 is tightly
regulated by systems-level feedbacks. Because Cdk1-Clb2 inhibits Sic1 by
phosphorylating Sic1 and making Sic1 available for degradation via
ubiquitylation, APC-Cdh1-dependent degradation of Cdk1-Clb2 not only
decreases the level of available Cdk1-Clb2 complexes but also increases
the level of Sic1 which in turn further inhibits the function of
Cdk1-Clb2.
This activation of the double negative feedback loop is initiated from
APC-Cdc20-dependent degradation of Cdk1-Clb2 and release of Cdc14 from
nucleolar protein Net1/Cfi1.
FEAR (Cdc14 early anaphase release) pathway facilitates
Clb2-Cdk1-dependent phosphorylation of Net1 which transiently releases
Cdc14 from Net1.
The released Cdc14 and Clb2-Cdk1 complexes go onto form spindles that
activates mitotic exit network (MEN). MEN allows sustained release of
Cdc14 from the nucleolus,
and Cdc14 counters the activity of Clb2-Cdk1 by activating Cdh1 and
stabilizing Sic1 through activation of Sic1-transcriptional activator
Swi5.
Sic1 positively regulates itself by inhibiting Cdk1-Clb2 to release
inhibition of Swi5, and Cdh1 also positively regulates itself by
inhibiting Clb2-Cdk1 to release inhibition of MEN which can activate
Cdc14 and subsequently Cdh1 itself. The double-negative feedback loop,
formed by APC-Cdh1 and Sic1, is required to maintain low Clb2-Cdk1
activity because Clb2 auto-activates its synthesis by activating
transcriptional factors, Fkh2–Mcm1 Ndd1 complex.
Implications
Eukaryotic
cell cycle consists of various checkpoints and feedback loops to ensure
faithful and successful cell division. During mitosis for example, when
duplicated chromosomes are improperly attached to mitotic spindle, spindle assembly checkpoint
(SAC) proteins including Mad and Bub inhibit APC-Cdc20 to delay entry
into anaphase and B-type cyclin degradations. In addition, when mitotic
spindles are misaligned, MEN and subsequently Cdc14 are inhibited in a
Bub2 and Bfa1-dependent manner to prevent degradation of mitotic cyclins
and anaphase entry.
Sic1 is a nice example demonstrating how systems-level feedbacks
interact to sense the environmental conditions and trigger cell cycle
transitions. Even though actual M-G1 transition is vastly complex with
numerous proteins and regulations involved, dynamical systems approach
allows simplification of this complex system into bistable input/output
relation with two saddle-node bifurcations in which the output (mitotic
exit) depends on critical concentration of Sic1. Using one-dimensional
analysis, it might be possible to explain many of the irreversible
transition points in the eukaryotic cell cycle that are governed by
systems-level control and feedback. Other examples of irreversible
transition points include Start (irreversible commitment to a new cell
division cycle) that can be explained by irreversible bistable switch
whose control parameter is tightly regulated by the systemic feedbacks
involving Cln2, Whi5, and SBF.