Inheritance
of dwarfing in maize. Demonstrating the heights of plants from the two
parent variations and their F1 heterozygous hybrid (centre)
Dominance, in genetics, is the phenomenon of one variant (allele) of a gene on a chromosome masking or overriding the effect of a different variant of the same gene on the other copy of the chromosome. The first variant is termed dominant and the second recessive. This state of having two different variants of the same gene on each chromosome is originally caused by a mutation in one of the genes, either new (de novo) or inherited. The terms autosomal dominant or autosomal recessive are used to describe gene variants on non-sex chromosomes (autosomes) and their associated traits, while those on sex chromosomes (allosomes) are termed X-linked dominant, X-linked recessive or Y-linked,
and these show a very different inheritance and presentation pattern to
autosomal traits which depends on the sex of the individual. Additionally, there are other forms of dominance such as incomplete dominance, in which a gene variant has a partial effect compared to when it is present on both chromosomes, and co-dominance, in which different variants on each chromosome both show their associated traits.
Dominance is not inherent to an allele or its traits (phenotype).
It is a strictly relative effect between two alleles of a given gene of
any function; one allele can be dominant over a second allele of the
same gene, recessive to a third and co-dominant with a fourth. Additionally, one allele may be dominant for one trait but not others.
Often, the dominant allele codes for a functional protein
whereas the recessive allele does not. Hence, many autosomal recessive
conditions are the result of lack of a functioning protein, for example an enzyme. On the other hand, many autosomal dominant conditions result in the presence of a diseased form of a critical protein or simply not enough expressivity of the critical protein.
Dominance is a key concept in Mendelian inheritance and classical genetics. Letters and Punnett squares
are used to demonstrate the principles of dominance in teaching, and
the use of upper case letters for dominant alleles and lower case
letters for recessive alleles is a widely followed convention. A classic
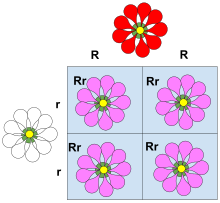
example of dominance is the inheritance of seed shape in peas. Peas may be round, associated with allele R, or wrinkled, associated with allele r. In this case, three combinations of alleles (genotypes) are possible: RR, Rr, and rr. The RR (homozygous) individuals have round peas, and the rr (homozygous) individuals have wrinkled peas. In Rr (heterozygous) individuals, the R allele masks the presence of the r allele, so these individuals also have round peas. Thus, allele R is dominant over allele r, and allele r is recessive to allele R.
Dominance differs from epistasis, the phenomenon of an allele of one gene masking the effect of alleles of a different gene.
Background
The concept of dominance was introduced by Gregor Johann Mendel.
Though Mendel, "The Father of Genetics", first used the term in the
1860s, it was not widely known until the early twentieth century. Mendel
observed that, for a variety of traits of garden peas having to do with
the appearance of seeds, seed pods, and plants, there were two discrete
phenotypes, such as round versus wrinkled seeds, yellow versus green
seeds, red versus white flowers or tall versus short plants. When bred
separately, the plants always produced the same phenotypes, generation
after generation. However, when lines with different phenotypes were
crossed (interbred), one and only one of the parental phenotypes showed
up in the offspring (green, or round, or red, or tall). However, when
these hybrid
plants were crossed, the offspring plants showed the two original
phenotypes, in a characteristic 3:1 ratio, the more common phenotype
being that of the parental hybrid plants. Mendel reasoned that each
parent in the first cross was a homozygote for different alleles (one
parent AA and the other parent aa), that each contributed one allele to
the offspring, with the result that all of these hybrids were
heterozygotes (Aa), and that one of the two alleles in the hybrid cross
dominated expression of the other: A masked a. The final cross between
two heterozygotes (Aa X Aa) would produce AA, Aa, and aa offspring in a
1:2:1 genotype ratio with the first two classes showing the (A)
phenotype, and the last showing the (a) phenotype, thereby producing the
3:1 phenotype ratio.
Mendel did not use the terms gene, allele, phenotype, genotype,
homozygote, and heterozygote, all of which were introduced later. He did
introduce the notation of capital and lowercase letters for dominant
and recessive alleles, respectively, still in use today.
Chromosomes, genes, and alleles
an autosomal dominant pattern.
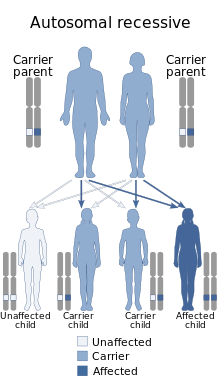
an autosomal recessive pattern.
Most animals and some plants have paired chromosomes, and are described as diploid. They have two versions of each chromosome, one contributed by the mother's ovum, and the other by the father's sperm, known as gametes, described as haploid, and created through meiosis. These gametes then fuse during fertilization during sexual reproduction, into a new single cell zygote,
which divides multiple times, resulting in a new organism with the same
number of pairs of chromosomes in each (non-gamete) cell as its
parents.
Each chromosome of a matching (homologous) pair is structurally similar to the other, and has a very similar DNA sequence (loci, singular locus). The DNA in each chromosome functions as a series of discrete genes
that influence various traits. Thus, each gene also has a corresponding
homologue, which may exist in different versions called alleles. The alleles at the same locus on the two homologous chromosomes may be identical or different.
The blood type of a human is determined by a gene that creates an A, B, AB or O
blood type and is located in the long arm of chromosome nine. There are
three different alleles that could be present at this locus, but only
two can be present in any individual, one inherited from their mother
and one from their father.
If two alleles of a given gene are identical, the organism is
called a homozygote and is said to be homozygous with respect to that
gene; if instead the two alleles are different, the organism is a
heterozygote and is heterozygous. The genetic makeup of an organism,
either at a single locus or over all its genes collectively, is called
its genotype.
The genotype of an organism directly and indirectly affects its
molecular, physical, and other traits, which individually or
collectively are called its phenotype. At heterozygous gene loci, the two alleles interact to produce the phenotype.
Dominance
Complete dominance
In
complete dominance, the effect of one allele in a heterozygous genotype
completely masks the effect of the other. The allele that masks the
other is said to be dominant to the latter, and the allele that is masked is said to be recessive to the former.
Complete dominance, therefore, means that the phenotype of the
heterozygote is indistinguishable from that of the dominant homozygote.
A classic example of dominance is the inheritance of seed shape (pea shape) in peas. Peas may be round (associated with allele R) or wrinkled (associated with allele r). In this case, three combinations of alleles (genotypes) are possible: RR and rr are homozygous and Rr is heterozygous. The RR individuals have round peas and the rr individuals have wrinkled peas. In Rr individuals the R allele masks the presence of the r allele, so these individuals also have round peas. Thus, allele R is completely dominant to allele r, and allele r is recessive to allele R.
Incomplete dominance:
This Punnett square illustrates incomplete dominance. In this example, the red petal trait associated with the R allele
recombines with the white petal trait of the r allele. The plant
incompletely expresses the dominant trait (R) causing plants with the Rr
genotype to express flowers with less red pigment resulting in pink
flowers. The colors are not blended together, the dominant trait is just
expressed less strongly.
Incomplete dominance (also called partial dominance, semi-dominance or intermediate inheritance)
occurs when the phenotype of the heterozygous genotype is distinct from
and often intermediate to the phenotypes of the homozygous genotypes.
For example, the snapdragon
flower color is homozygous for either red or white. When the red
homozygous flower is paired with the white homozygous flower, the result
yields a pink snapdragon flower. The pink snapdragon is the result of
incomplete dominance. A similar type of incomplete dominance is found in
the four o'clock plant wherein pink color is produced when true-bred parents of white and red flowers are crossed. In quantitative genetics,
where phenotypes are measured and treated numerically, if a
heterozygote's phenotype is exactly between (numerically) that of the
two homozygotes, the phenotype is said to exhibit no dominance at all, i.e. dominance exists only when the heterozygote's phenotype measure lies closer to one homozygote than the other.
When plants of the F1 generation are self-pollinated, the phenotypic and genotypic ratio of the F2 generation will be 1:2:1 (Red:Pink:White).
Co-dominance
Co-dominance in a Camellia cultivar
A and B blood types in humans show co-dominance, but the O type is recessive to A and B.
This
diagram shows co-dominance. In this example a white bull (WW) mates
with a red cow (RR), and their offspring exhibit co-dominance expressing
both white and red hairs.
Co-dominance occurs when the contributions of both alleles are visible in the phenotype.
For example, in the ABO blood group system, chemical modifications to a glycoprotein (the H antigen) on the surfaces of blood cells are controlled by three alleles, two of which are co-dominant to each other (IA, IB) and dominant over the recessive i at the ABO locus. The IA and IB alleles produce different modifications. The enzyme coded for by IA adds an N-acetylgalactosamine to the membrane-bound H antigen. The IB enzyme adds a galactose. The i allele produces no modification. Thus IA and IB alleles are each dominant to i (IAIA and IAi individuals both have type A blood, and IBIB and IBi individuals both have type B blood, but IAIB individuals have both modifications on their blood cells and thus have type AB blood, so the IA and IB alleles are said to be co-dominant).
Another example occurs at the locus for the beta-globin component of hemoglobin, where the three molecular phenotypes of HbA/HbA, HbA/HbS, and HbS/HbS are all distinguishable by protein electrophoresis. (The medical condition produced by the heterozygous genotype is called sickle-cell trait and is a milder condition distinguishable from sickle-cell anemia, thus the alleles show incomplete dominance
with respect to anemia, see above). For most gene loci at the molecular
level, both alleles are expressed co-dominantly, because both are transcribed into RNA.
Co-dominance, where allelic products co-exist in the phenotype,
is different from incomplete dominance, where the quantitative
interaction of allele products produces an intermediate phenotype. For
example, in co-dominance, a red homozygous flower and a white homozygous
flower will produce offspring that have red and white spots. When
plants of the F1 generation are self-pollinated, the phenotypic and
genotypic ratio of the F2 generation will be 1:2:1 (Red:Spotted:White).
These ratios are the same as those for incomplete dominance. Again, this
classical terminology is inappropriate – in reality such cases should
not be said to exhibit dominance at all.
Addressing common misconceptions
While it is often convenient to talk about a recessive allele or a dominant trait,
dominance is not inherent to either an allele or its phenotype.
Dominance is a relationship between two alleles of a gene and their
associated phenotypes. A "dominant" allele is dominant to a particular
allele of the same gene that can be inferred from the context, but it
may be recessive to a third allele, and codominant to a fourth.
Similarly, a "recessive" trait is a trait associated with a particular
recessive allele implied by the context, but that same trait may occur
in a different context where it is due to some other gene and a dominant
allele.
Dominance is unrelated to the nature of the phenotype itself,
that is, whether it is regarded as "normal" or "abnormal," "standard" or
"nonstandard," "healthy" or "diseased," "stronger" or "weaker," or more
or less extreme. A dominant or recessive allele may account for any of
these trait types.
Dominance does not determine whether an allele is deleterious, neutral or advantageous. However, selection
must operate on genes indirectly through phenotypes, and dominance
affects the exposure of alleles in phenotypes, and hence the rate of
change in allele frequencies under selection. Deleterious recessive
alleles may persist in a population at low frequencies, with most copies
carried in heterozygotes, at no cost to those individuals. These rare
recessives are the basis for many hereditary genetic disorders.
Dominance is also unrelated to the distribution of alleles in the
population. Both dominant and recessive alleles can be extremely common
or extremely rare.
Nomenclature
In genetics, symbols began as algebraic placeholders. When one allele
is dominant to another, the oldest convention is to symbolize the
dominant allele with a capital letter. The recessive allele is assigned
the same letter in lower case. In the pea example, once the dominance
relationship between the two alleles is known, it is possible to
designate the dominant allele that produces a round shape by a
capital-letter symbol R, and the recessive allele that produces a wrinkled shape by a lower-case symbol r. The homozygous dominant, heterozygous, and homozygous recessive genotypes are then written RR, Rr, and rr, respectively. It would also be possible to designate the two alleles as W and w, and the three genotypes WW, Ww, and ww, the first two of which produced round peas and the third wrinkled peas. The choice of "R" or "W"
as the symbol for the dominant allele does not pre-judge whether the
allele causing the "round" or "wrinkled" phenotype when homozygous is
the dominant one.
A gene may have several alleles. Each allele is symbolized by the
locus symbol followed by a unique superscript. In many species, the
most common allele in the wild population is designated the wild type
allele. It is symbolized with a + character as a superscript. Other
alleles are dominant or recessive to the wild type allele. For recessive
alleles, the locus symbol is in lower case letters. For alleles with
any degree of dominance to the wild type allele, the first letter of the
locus symbol is in upper case. For example, here are some of the
alleles at the a locus of the laboratory mouse, Mus musculus: Ay, dominant yellow; a+, wild type; and abt, black and tan. The abt allele is recessive to the wild type allele, and the Ay allele is codominant to the wild type allele. The Ay allele is also codominant to the abt allele, but showing that relationship is beyond the limits of the rules for mouse genetic nomenclature.
Rules of genetic nomenclature have evolved as genetics has become
more complex. Committees have standardized the rules for some species,
but not for all. Rules for one species may differ somewhat from the
rules for a different species.
Relationship to other genetic concepts
Multiple alleles
Although any individual of a diploid organism has at most two different alleles at any one locus (barring aneuploidies),
most genes exist in a large number of allelic versions in the
population as a whole. If the alleles have different effects on the
phenotype, sometimes their dominance relationships can be described as a
series.
For example, coat color in domestic cats is affected by a series of alleles of the TYR gene (which encodes the enzyme tyrosinase). The alleles C, cb, cs, and ca (full colour, Burmese, Siamese, and albino, respectively) produce different levels of pigment and hence different levels of colour dilution. The C allele (full colour) is completely dominant over the last three and the ca allele (albino) is completely recessive to the first three.
Autosomal versus sex-linked dominance
In humans and other mammal species, sex is determined by two sex chromosomes called the X chromosome and the Y chromosome. Human females are typically XX; males are typically XY. The remaining pairs of chromosome are found in both sexes and are called autosomes;
genetic traits due to loci on these chromosomes are described as
autosomal, and may be dominant or recessive. Genetic traits on the X and Y
chromosomes are called sex-linked, because they are linked to sex
chromosomes, not because they are characteristic of one sex or the
other. In practice, the term almost always refers to X-linked
traits and a great many such traits (such as red-green colour vision
deficiency) are not affected by sex. Females have two copies of every
gene locus found on the X chromosome, just as for the autosomes, and the
same dominance relationships apply. Males, however, have only one copy
of each X chromosome gene locus, and are described as hemizygous for these genes. The Y chromosome is much smaller than the X, and contains a much smaller set of genes, including, but not limited to, those that influence 'maleness', such as the SRY gene for testis determining factor.
Dominance rules for sex-linked gene loci are determined by their
behavior in the female: because the male has only one allele (except in
the case of certain types of Y chromosome aneuploidy),
that allele is always expressed regardless of whether it is dominant or
recessive.
Birds have oppositely sex chromosomes: male birds have ZZ and female
birds ZW chromosomes. However, inheritance of traits reminds XY-system
otherwise; male zebra finches may carry white colouring gene in their
one of two Z chromosome, but females develop white colouring always.
Grasshoppers have XO-system. Females have XX, but males only X. There is
no Y chromosome at all.
Epistasis
Epistasis ["epi + stasis = to sit on top"] is an interaction between alleles at two different gene loci that affect a single trait, which may sometimes resemble a dominance interaction between two different alleles at the same locus. Epistasis modifies the characteristic 9:3:3:1
ratio expected for two non-epistatic genes. For two loci, 14 classes of
epistatic interactions are recognized. As an example of recessive epistasis, one gene locus may determine whether a flower pigment is yellow (AA or Aa) or green (aa), while another locus determines whether the pigment is produced (BB or Bb) or not (bb). In a bb plant, the flowers will be white, irrespective of the genotype of the other locus as AA, Aa, or aa. The bb combination is not dominant to the A allele: rather, the B gene shows recessive epistasis to the A gene, because the B locus when homozygous for the recessive allele (bb) suppresses phenotypic expression of the A locus. In a cross between two AaBb plants, this produces a characteristic 9:3:4 ratio, in this case of yellow : green : white flowers.
In dominant epistasis, one gene locus may determine yellow or green pigment as in the previous example: AA and Aa are yellow, and aa are green. A second locus determines whether a pigment precursor is produced (dd) or not (DD or Dd). Here, in a DD or Dd plant, the flowers will be colorless irrespective of the genotype at the A locus, because of the epistatic effect of the dominant D allele. Thus, in a cross between two AaDd plants, 3/4 of the plants will be colorless, and the yellow and green phenotypes are expressed only in dd plants. This produces a characteristic 12:3:1 ratio of white : yellow : green plants.
Supplementary epistasis occurs when two loci affect the same phenotype. For example, if pigment color is produced by CC or Cc but not cc, and by DD or Dd but not dd, then pigment is not produced in any genotypic combination with either cc or dd. That is, both loci must have at least one dominant allele to produce the phenotype. This produces a characteristic 9:7 ratio of pigmented to unpigmented plants. Complementary epistasis in contrast produces an unpigmented plant if and only if the genotype is cc and dd, and the characteristic ratio is 15:1 between pigmented and unpigmented plants.
Classical genetics considered epistatic interactions between two
genes at a time. It is now evident from molecular genetics that all gene
loci are involved in complex interactions with many other genes (e.g.,
metabolic pathways may involve scores of genes), and that this creates
epistatic interactions that are much more complex than the classic
two-locus models.
Hardy–Weinberg principle (estimation of carrier frequency)
The frequency of the heterozygous state (which is the carrier state for a recessive trait) can be estimated using the Hardy–Weinberg formula:
This formula applies to a gene with exactly two alleles and
relates the frequencies of those alleles in a large population to the
frequencies of their three genotypes in that population.
For example, if p is the frequency of allele A, and q is the frequency of allele a then the terms p2, 2pq, and q2 are the frequencies of the genotypes AA, Aa and aa respectively. Since the gene has only two alleles, all alleles must be either A or a and p + q = 1. Now, if A is completely dominant to a then the frequency of the carrier genotype Aa cannot be directly observed (since it has the same traits as the homozygous genotype AA),
however it can be estimated from the frequency of the recessive trait
in the population, since this is the same as that of the homozygous
genotype aa. i.e. the individual allele frequencies can be estimated: q = √f(aa), p = 1 − q, and from those the frequency of the carrier genotype can be derived: f(Aa) = 2pq.
This formula relies on a number of assumptions
and an accurate estimate of the frequency of the recessive trait. In
general, any real-world situation will deviate from these assumptions to
some degree, introducing corresponding inaccuracies into the estimate.
If the recessive trait is rare, then it will be hard to estimate its
frequency accurately, as a very large sample size will be needed.
Dominant versus advantageous
The
property of "dominant" is sometimes confused with the concept of
advantageous and the property of "recessive" is sometimes confused with
the concept of deleterious, but the phenomena are distinct. Dominance
describes the phenotype of heterozygotes with regard to the phenotypes
of the homozygotes and without respect to the degree to which different
phenotypes may be beneficial or deleterious. Since many genetic disease
alleles are recessive and because the word dominance has a positive
connotation, the assumption that the dominant phenotype is superior with
respect to fitness is often made. This is not assured however; as
discussed below while most genetic disease alleles are deleterious and
recessive, not all genetic diseases are recessive.
Nevertheless, this confusion has been pervasive throughout the
history of genetics and persists to this day. Addressing this confusion
was one of the prime motivations for the publication of the Hardy-Weinberg principle.
Molecular mechanisms
The
molecular basis of dominance was unknown to Mendel. It is now
understood that a gene locus includes a long series (hundreds to
thousands) of bases or nucleotides of deoxyribonucleic acid (DNA) at a particular point on a chromosome. The central dogma of molecular biology states that "DNA makes RNA makes protein", that is, that DNA is transcribed to make an RNA copy, and RNA is translated
to make a protein. In this process, different alleles at a locus may or
may not be transcribed, and if transcribed may be translated to
slightly different versions of the same protein (called isoforms). Proteins often function as enzymes
that catalyze chemical reactions in the cell, which directly or
indirectly produce phenotypes. In any diploid organism, the DNA
sequences of the two alleles present at any gene locus may be identical
(homozygous) or different (heterozygous). Even if the gene locus is
heterozygous at the level of the DNA sequence, the proteins made by each
allele may be identical. In the absence of any difference between the
protein products, neither allele can be said to be dominant (see co-dominance, above). Even if the two protein products are slightly different (allozymes),
it is likely that they produce the same phenotype with respect to
enzyme action, and again neither allele can be said to be dominant.
Loss of function and haplosufficiency
Dominance
typically occurs when one of the two alleles is non-functional at the
molecular level, that is, it is not transcribed or else does not produce
a functional protein product. This can be the result of a mutation that alters the DNA sequence of the allele.
An organism homozygous for the non-functional allele will generally
show a distinctive phenotype, due to the absence of the protein product.
For example, in humans and other organisms, the unpigmented skin of the
albino phenotype
results when an individual is homozygous for an allele that encodes a
non-functional version of an enzyme needed to produce the skin pigment melanin.
It is important to understand that it is not the lack of function that
allows the allele to be described as recessive: this is the interaction
with the alternative allele in the heterozygote. Three general types of
interaction are possible:
- In the typical case, the single functional allele makes sufficient protein to produce a phenotype identical to that of the homozygote: this is called haplosufficiency. For example, suppose the standard amount of enzyme produced in the functional homozygote is 100%, with the two functional alleles contributing 50% each. The single functional allele in the heterozygote produces 50% of the standard amount of enzyme, which is sufficient to produce the standard phenotype. If the heterozygote and the functional-allele homozygote have identical phenotypes, the functional allele is dominant to the non-functional allele. This occurs at the albino gene locus: the heterozygote produces sufficient enzyme to convert the pigment precursor to melanin, and the individual has standard pigmentation.
- Less commonly, the presence of a single functional allele gives a phenotype that is not normal but less severe than that of the non-functional homozygote. This occurs when the functional allele is not haplo-sufficient. The terms haplo-insufficiency and incomplete dominance are typically applied to these cases. The intermediate interaction occurs where the heterozygous genotype produces a phenotype intermediate between the two homozygotes. Depending on which of the two homozygotes the heterozygote most resembles, one allele is said to show incomplete dominance over the other. For example, in humans the Hb gene locus is responsible for the Beta-chain protein (HBB) that is one of the two globin proteins that make up the blood pigment hemoglobin. Many people are homozygous for an allele called HbA; some persons carry an alternative allele called HbS, either as homozygotes or heterozygotes. The hemoglobin molecules of HbS/HbS homozygotes undergo a change in shape that distorts the morphology of the red blood cells, and causes a severe, life-threatening form of anemia called sickle-cell anemia. Persons heterozygous HbA/HbS for this allele have a much less severe form of anemia called sickle-cell trait. Because the disease phenotype of HbA/HbS heterozygotes is more similar to but not identical to the HbA/HbA homozygote, the HbA allele is said to be incompletely dominant to the HbS allele.
- Rarely, a single functional allele in the heterozygote may produce insufficient gene product for any function of the gene, and the phenotype resembles that of the homozygote for the non-functional allele. This complete haploinsufficiency is very unusual. In these cases, the non-functional allele would be said to be dominant to the functional allele. This situation may occur when the non-functional allele produces a defective protein that interferes with the proper function of the protein produced by the standard allele. The presence of the defective protein "dominates" the standard protein, and the disease phenotype of the heterozygote more closely resembles that of the homozygote for two defective alleles. The term "dominant" is often incorrectly applied to defective alleles whose homozygous phenotype has not been examined, but which cause a distinct phenotype when heterozygous with the normal allele. This phenomenon occurs in a number of trinucleotide repeat diseases, one example being Huntington's disease.
Dominant-negative mutations
Many
proteins are normally active in the form of a multimer, an aggregate of
multiple copies of the same protein, otherwise known as a homomultimeric protein or homooligomeric protein. In fact, a majority of the 83,000 different enzymes from 9800 different organisms in the BRENDA Enzyme Database represent homooligomers.
When the wild-type version of the protein is present along with a
mutant version, a mixed multimer can be formed. A mutation that leads to
a mutant protein that disrupts the activity of the wild-type protein in
the multimer is a dominant-negative mutation.
A dominant-negative mutation may arise in a human somatic cell
and provide a proliferative advantage to the mutant cell, leading to its
clonal expansion. For instance, a dominant-negative mutation in a gene
necessary for the normal process of programmed cell death (Apoptosis)
in response to DNA damage can make the cell resistant to apoptosis.
This will allow proliferation of the clone even when excessive DNA
damage is present. Such dominant-negative mutations occur in the tumor
suppressor gene p53. The P53 wild-type protein is normally present as a four-protein multimer (oligotetramer). Dominant-negative p53
mutations occur in a number of different types of cancer and
pre-cancerous lesions (e.g. brain tumors, breast cancer, oral
pre-cancerous lesions and oral cancer).
Dominant-negative mutations also occur in other tumor suppressor
genes. For instance two dominant-negative germ line mutations were
identified in the Ataxia telangiectasia mutated (ATM) gene which increases susceptibility to breast cancer. Dominant negative mutations of the transcription factor C/EBPα can cause acute myeloid leukemia. Inherited dominant negative mutations can also increase the risk of diseases other than cancer. Dominant-negative mutations in Peroxisome proliferator-activated receptor gamma (PPARγ) are associated with severe insulin resistance, diabetes mellitus and hypertension.
Dominant-negative mutations have also been described in organisms other than humans. In fact, the first study reporting a mutant protein
inhibiting the normal function of a wild-type protein in a mixed
multimer was with the bacteriophage T4 tail fiber protein GP37.
Mutations that produce a truncated protein rather than a full-length
mutant protein seem to have the strongest dominant-negative effect in
the studies of P53, ATM, C/EBPα, and bacteriophage T4 GP37.
Dominant and recessive genetic diseases in humans
In
humans, many genetic traits or diseases are classified simply as
"dominant" or "recessive". Especially with so-called recessive diseases,
which are indeed a factor of recessive genes, but can oversimplify the
underlying molecular basis and lead to misunderstanding of the nature of
dominance. For example, the recessive genetic disease phenylketonuria (PKU) results from any of a large number (>60) of alleles at the gene locus for the enzyme phenylalanine hydroxylase (PAH). Many of these alleles produce little or no PAH, as a result of which the substrate phenylalanine (Phe) and its metabolic byproducts accumulate in the central nervous system and can cause severe intellectual disability if untreated.
To illustrate these nuances, the genotypes and phenotypic
consequences of interactions among three hypothetical PAH alleles are
shown in the following table:
| Genotype | PAH activity | [Phe] conc | PKU ? |
|---|---|---|---|
| AA | 100% | 60 uM | No |
| AB | 30% | 120 uM | No |
| CC | 5% | 200 ~ 300 uM | Hyperphenylalaninemia |
| BB | 0.3% | 600 ~ 2400 uM | Yes |
In unaffected persons homozygous for a standard functional allele (AA), PAH activity is standard (100%), and the concentration of phenylalanine in the blood [Phe] is about 60 uM. In untreated persons homozygous for one of the PKU alleles (BB), PAH activity is close to zero, [Phe] ten to forty times standard, and the individual manifests PKU.
In the AB heterozygote, PAH activity is only 30% (not 50%) of standard, blood [Phe] is elevated two-fold, and the person does not manifest PKU. Thus, the A allele is dominant to the B allele with respect to PKU, but the B allele is incompletely dominant to the A allele with respect to its molecular effect, determination of PAH activity level (0.3% < 30% << 100%). Finally, the A allele is an incomplete dominant to B
with respect to [Phe], as 60 uM < 120 uM << 600 uM. Note once
more that it is irrelevant to the question of dominance that the
recessive allele produces a more extreme [Phe] phenotype.
For a third allele C, a CC homozygote produces a very small amount of PAH enzyme, which results in a somewhat elevated level of [Phe] in the blood, a condition called hyperphenylalaninemia, which does not result in intellectual disability.
That is, the dominance relationships of any two alleles may vary
according to which aspect of the phenotype is under consideration. It is
typically more useful to talk about the phenotypic consequences of the
allelic interactions involved in any genotype, rather than to try to
force them into dominant and recessive categories.