Coral reefs are diverse underwater ecosystems held together by calcium carbonate structures secreted by corals. Coral reefs are built by colonies of tiny animals found in marine water that contain few nutrients. Most coral reefs are built from stony corals, which in turn consist of polyps that cluster in groups. The polyps belong to a group of animals known as Cnidaria, which also includes sea anemones and jellyfish. Unlike sea anemones, corals secrete hard carbonate exoskeletons which support and protect the coral polyps. Most reefs grow best in warm, shallow, clear, sunny and agitated water.
Often called "rainforests of the sea", shallow coral reefs form some of the most diverse ecosystems on Earth. They occupy less than 0.1% of the world's ocean surface, about half the area of France, yet they provide a home for at least 25% of all marine species, including fish, mollusks, worms, crustaceans, echinoderms, sponges, tunicates and other cnidarians. Paradoxically, coral reefs flourish even though they are surrounded by ocean waters that provide few nutrients. They are most commonly found at shallow depths in tropical waters, but deep water and cold water corals also exist on smaller scales in other areas.
Coral reefs deliver ecosystem services to tourism, fisheries and shoreline protection. The annual global economic value of coral reefs is estimated between US$30–375 billion. However, coral reefs are fragile ecosystems, partly because they are very sensitive to water temperature. They are under threat from climate change, oceanic acidification, blast fishing, cyanide fishing for aquarium fish, sunscreen use, overuse of reef resources, and harmful land-use practices, including urban and agricultural runoff and water pollution, which can harm reefs by encouraging excess algal growth.
Formation
Most of the coral reefs we can see today were formed after the last glacial period when melting ice caused the sea level to rise and flood the continental shelves. This means that most modern coral reefs are less than 10,000 years old. As communities established themselves on the shelves, the reefs grew upwards, pacing rising sea levels. Reefs that rose too slowly could become drowned reefs. They are covered by so much water that there was insufficient light. Coral reefs are found in the deep sea away from continental shelves, around oceanic islands and as atolls. The vast majority of these islands are volcanic in origin. The few exceptions have tectonic origins where plate movements have lifted the deep ocean floor on the surface.
In 1842 in his first monograph, The Structure and Distribution of Coral Reefs, Charles Darwin set out his theory of the formation of atoll reefs, an idea he conceived during the voyage of the Beagle. He theorized uplift and subsidence of the Earth's crust under the oceans formed the atolls. Darwin’s theory sets out a sequence of three stages in atoll formation. It starts with a fringing reef forming around an extinct volcanic island
as the island and ocean floor subsides. As the subsidence continues,
the fringing reef becomes a barrier reef, and ultimately an atoll reef.
-
Darwin’s theory starts with a volcanic island which becomes extinct
-
As the island and ocean floor subside, coral growth builds a fringing reef, often including a shallow lagoon between the land and the main reef.
-
As the subsidence continues, the fringing reef becomes a larger barrier reef further from the shore with a bigger and deeper lagoon inside.
-
Ultimately, the island sinks below the sea, and the barrier reef becomes an atoll enclosing an open lagoon.
Where the bottom is rising, fringing reefs can grow around the coast, but coral raised above sea level dies and becomes white limestone. If the land subsides slowly, the fringing reefs keep pace by growing upwards on a base of older, dead coral, forming a barrier reef enclosing a lagoon between the reef and the land. A barrier reef can encircle an island, and once the island sinks below sea level a roughly circular atoll of growing coral continues to keep up with the sea level, forming a central lagoon. Barrier reefs and atolls do not usually form complete circles, but are broken in places by storms. Like sea level rise, a rapidly subsiding bottom can overwhelm coral growth, killing the coral polyps and the reef, due to what is called coral drowning. Corals that rely on zooxanthellae can drown when the water becomes too deep for their symbionts to adequately photosynthesize, due to decreased light exposure.
The two main variables determining the geomorphology, or shape, of coral reefs are the nature of the underlying substrate on which they rest, and the history of the change in sea level relative to that substrate.
The approximately 20,000-year-old Great Barrier Reef offers an example of how coral reefs formed on continental shelves. Sea level was then 120 m (390 ft) lower than in the 21st century. As sea level rose, the water and the corals encroached on what had been hills of the Australian coastal plain. By 13,000 years ago, sea level had risen to 60 m (200 ft) lower than at present, and many hills of the coastal plains had become continental islands. As the sea level rise continued, water topped most of the continental islands. The corals could then overgrow the hills, forming the present cays and reefs. Sea level on the Great Barrier Reef has not changed significantly in the last 6,000 years, and the age of the modern living reef structure is estimated to be between 6,000 and 8,000 years. Although the Great Barrier Reef formed along a continental shelf, and not around a volcanic island, Darwin's principles apply. Development stopped at the barrier reef stage, since Australia is not about to submerge. It formed the world's largest barrier reef, 300–1,000 m (980–3,280 ft) from shore, stretching for 2,000 km (1,200 mi).
Healthy tropical coral reefs grow horizontally from 1 to 3 cm (0.39 to 1.18 in) per year, and grow vertically anywhere from 1 to 25 cm (0.39 to 9.84 in) per year; however, they grow only at depths shallower than 150 m (490 ft) because of their need for sunlight, and cannot grow above sea level.
Materials
As the name implies, the bulk of coral reefs is made up of coral skeletons from mostly intact coral colonies. As other chemical elements present in corals become incorporated into the calcium carbonate deposits, aragonite is formed. However, shell fragments and the remains of calcareous algae such as the green-segmented genus Halimeda can add to the reef's ability to withstand damage from storms and other threats. Such mixtures are visible in structures such as Eniwetok Atoll.Types
The three principal reef types are:- Fringing reef – directly attached to a shore, or borders it with an intervening shallow channel or lagoon
- Barrier reef – reef separated from a mainland or island shore by a deep channel or lagoon
- Atoll reef – more or less circular or continuous barrier reef extends all the way around a lagoon without a central island
- Patch reef – common, isolated, comparatively small reef outcrop, usually within a lagoon or embayment, often circular and surrounded by sand or seagrass
- Apron reef – short reef resembling a fringing reef, but more sloped; extending out and downward from a point or peninsular shore
- Bank reef – linear or semicircular shaped-outline, larger than a patch reef
- Ribbon reef – long, narrow, possibly winding reef, usually associated with an atoll lagoon
- Table reef – isolated reef, approaching an atoll type, but without a lagoon
- Habili – reef specific to the Red Sea; does not reach the surface near enough to cause visible surf; may be a hazard to ships (from the Arabic for "unborn")
- Microatoll – community of species of corals; vertical growth limited by average tidal height; growth morphologies offer a low-resolution record of patterns of sea level change; fossilized remains can be dated using radioactive carbon dating and have been used to reconstruct Holocene sea levels
- Cays – small, low-elevation, sandy islands formed on the surface of coral reefs from eroded material that piles up, forming an area above sea level; can be stabilized by plants to become habitable; occur in tropical environments throughout the Pacific, Atlantic and Indian Oceans (including the Caribbean and on the Great Barrier Reef and Belize Barrier Reef), where they provide habitable and agricultural land
- Seamount or guyot – formed when a coral reef on a volcanic island subsides; tops of seamounts are rounded and guyots are flat; flat tops of guyots, or tablemounts, are due to erosion by waves, winds, and atmospheric processes
Zones
Coral reef ecosystems contain distinct zones that represent different kinds of habitats. Usually, three major zones are recognized: the fore reef, reef crest, and the back reef (frequently referred to as the reef lagoon).
All three zones are physically and ecologically interconnected. Reef life and oceanic processes create opportunities for exchange of seawater, sediments, nutrients, and marine life among one another.
Thus, they are integrated components of the coral reef ecosystem, each playing a role in the support of the reefs' abundant and diverse fish assemblages.
Most coral reefs exist in shallow waters less than 50 m deep. Some inhabit tropical continental shelves where cool, nutrient rich upwelling does not occur, such as Great Barrier Reef. Others are found in the deep ocean surrounding islands or as atolls, such as in the Maldives. The reefs surrounding islands form when islands subside into the ocean, and atolls form when an island subsides below the surface of the sea.
Alternatively, Moyle and Cech distinguish six zones, though most reefs possess only some of the zones.
The reef surface is the shallowest part of the reef. It is subject to the surge and the rise and fall of tides. When waves pass over shallow areas, they shoal, as shown in the diagram at the right. This means the water is often agitated. These are the precise condition under which corals flourish. Shallowness means there is plenty of light for photosynthesis by the symbiotic zooxanthellae, and agitated water promotes the ability of coral to feed on plankton. However, other organisms must be able to withstand the robust conditions to flourish in this zone.
The off-reef floor is the shallow sea floor surrounding a reef. This zone occurs by reefs on continental shelves. Reefs around tropical islands and atolls drop abruptly to great depths, and do not have a floor. Usually sandy, the floor often supports seagrass meadows which are important foraging areas for reef fish.
The reef drop-off is, for its first 50 m, habitat for many reef fish who find shelter on the cliff face and plankton in the water nearby. The drop-off zone applies mainly to the reefs surrounding oceanic islands and atolls.
The reef face is the zone above the reef floor or the reef drop-off. This zone is often the most diverse area of the reef. Coral and calcareous algae growths provide complex habitats and areas which offer protection, such as cracks and crevices. Invertebrates and epiphytic algae provide much of the food for other organisms. A common feature on this forereef zone is spur and groove formations which serve to transport sediment downslope.
The reef flat is the sandy-bottomed flat, which can be behind the main reef, containing chunks of coral. This zone may border a lagoon and serve as a protective area, or it may lie between the reef and the shore, and in this case is a flat, rocky area. Fishes tend to prefer living in that flat, rocky area, compared to any other zone, when it is present.
The reef lagoon is an entirely enclosed region, which creates an area less affected by wave action that often contains small reef patches.
However, the "topography of coral reefs is constantly changing. Each reef is made up of irregular patches of algae, sessile invertebrates, and bare rock and sand. The size, shape and relative abundance of these patches changes from year to year in response to the various factors that favor one type of patch over another. Growing coral, for example, produces constant change in the fine structure of reefs. On a larger scale, tropical storms may knock out large sections of reef and cause boulders on sandy areas to move."
Locations
Coral reefs are estimated to cover 284,300 km2 (109,800 sq mi), just under 0.1% of the oceans' surface area. The Indo-Pacific region (including the Red Sea, Indian Ocean, Southeast Asia and the Pacific) account for 91.9% of this total. Southeast Asia accounts for 32.3% of that figure, while the Pacific including Australia accounts for 40.8%. Atlantic and Caribbean coral reefs account for 7.6%.
Although corals exist both in temperate and tropical waters, shallow-water reefs form only in a zone extending from approximately 30° N to 30° S of the equator. Tropical corals do not grow at depths of over 50 meters (160 ft). The optimum temperature for most coral reefs is 26–27 °C (79–81 °F), and few reefs exist in waters below 18 °C (64 °F). However, reefs in the Persian Gulf have adapted to temperatures of 13 °C (55 °F) in winter and 38 °C (100 °F) in summer. There are 37 species of scleractinian corals identified in such harsh environment around Larak Island.
Deep-water coral can exist at greater depths and colder temperatures at much higher latitudes, as far north as Norway. Although deep water corals can form reefs, very little is known about them.
Coral reefs are rare along the west coasts of the Americas and Africa, due primarily to upwelling and strong cold coastal currents that reduce water temperatures in these areas (respectively the Peru, Benguela and Canary streams). Corals are seldom found along the coastline of South Asia—from the eastern tip of India (Chennai) to the Bangladesh and Myanmar borders—as well as along the coasts of northeastern South America and Bangladesh, due to the freshwater release from the Amazon and Ganges Rivers respectively.
- The Great Barrier Reef—largest, comprising over 2,900 individual reefs and 900 islands stretching for over 2,600 kilometers (1,600 mi) off Queensland, Australia
- The Mesoamerican Barrier Reef System—second largest, stretching 1,000 kilometers (620 mi) from Isla Contoy at the tip of the Yucatán Peninsula down to the Bay Islands of Honduras
- The New Caledonia Barrier Reef—second longest double barrier reef, covering 1,500 kilometers (930 mi)
- The Andros, Bahamas Barrier Reef—third largest, following the east coast of Andros Island, Bahamas, between Andros and Nassau
- The Red Sea—includes 6000-year-old fringing reefs located around a 2,000 km (1,240 mi) coastline
- The Florida Reef Tract—largest continental US reef and the third largest coral barrier reef system in the world, extends from Soldier Key, located in Biscayne Bay, to the Dry Tortugas in the Gulf of Mexico
- Pulley Ridge—deepest photosynthetic coral reef, Florida
- Numerous reefs scattered over the Maldives
- The Philippines coral reef area, the second largest in Southeast Asia, is estimated at 26,000 square kilometers and holds an extraordinary diversity of species. Scientists have identified 915 reef fish species and more than 400 scleractinian coral species, 12 of which are endemic.
- The Raja Ampat Islands in Indonesia's West Papua province offer the highest known marine diversity.
- Bermuda is known for its northernmost coral reef system, located at 32.4° N and 64.8° W. The presence of coral reefs at this high latitude is due to the proximity of the Gulf Stream. Bermuda has a fairly consistent diversity of coral species, representing a subset of those found in the greater Caribbean.
- The world's northernmost individual coral reef so far discovered is located within a bay of Japan's Tsushima Island in the Korea Strait.
- The world's southernmost coral reef is at Lord Howe Island, in the Pacific Ocean off the east coast of Australia.
Biology
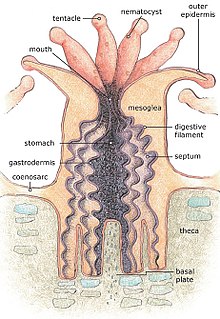
Alive corals are colonies of small animals embedded in calcium carbonate
shells. It is a mistake to think of coral as plants or rocks. Coral
heads consist of accumulations of individual animals called polyps, arranged in diverse shapes. Polyps are usually tiny, but they can range in size from a pinhead to 12 inches (30 cm) across.
Reef-building or hermatypic corals live only in the photic zone
(above 50 m), the depth to which sufficient sunlight penetrates the
water, allowing photosynthesis to occur. Coral polyps do not
photosynthesize, but have a symbiotic relationship with microscopic algae of the genus Symbiodinium,
commonly referred to as zooxanthellae. These organisms live within the
tissues of polyps and provide organic nutrients that nourish the polyp.
Because of this relationship, coral reefs grow much faster in clear
water, which admits more sunlight. Without their symbionts, coral growth
would be too slow to form significant reef structures. Corals get up to
90% of their nutrients from their symbionts.
Reefs grow as polyps and other organisms deposit calcium carbonate, the basis of coral, as a skeletal structure beneath and around themselves, pushing the coral head's top upwards and outwards. Waves, grazing fish (such as parrotfish), sea urchins, sponges, and other forces and organisms act as bioeroders,
breaking down coral skeletons into fragments that settle into spaces in
the reef structure or form sandy bottoms in associated reef lagoons.
Many other organisms living in the reef community contribute skeletal
calcium carbonate in the same manner. Coralline algae
are important contributors to reef structure in those parts of the reef
subjected to the greatest forces by waves (such as the reef front
facing the open ocean). These algae strengthen the reef structure by
depositing limestone in sheets over the reef surface.
Typical shapes for coral species are wrinkled brains, cabbages, table tops, antlers, wire strands and pillars. These shapes can depend on the life history of the coral, like light exposure and wave action, and events such as breakages.
Corals reproduce both sexually and asexually. An individual polyp uses both reproductive modes within its lifetime. Corals reproduce sexually by either internal or external fertilization. The reproductive cells are found on the mesenteries, membranes that radiate inward from the layer of tissue that lines the stomach cavity. Some mature adult corals are hermaphroditic; others are exclusively male or female. A few species change sex as they grow.
Internally fertilized eggs develop in the polyp for a period ranging from days to weeks. Subsequent development produces a tiny larva, known as a planula. Externally fertilized eggs develop during synchronized spawning. Polyps release eggs and sperm into the water en masse, simultaneously. Eggs disperse over a large area. The timing of spawning depends on time of year, water temperature, and tidal and lunar cycles. Spawning is most successful when there is little variation between high and low tide. The less water movement, the better the chance for fertilization. Ideal timing occurs in the spring. Release of eggs or planula usually occurs at night, and is sometimes in phase with the lunar cycle (three to six days after a full moon). The period from release to settlement lasts only a few days, but some planulae can survive afloat for several weeks. They are vulnerable to predation and environmental conditions. The lucky few planulae which successfully attach to substrate next confront competition for food and space.
There are eight clades of Symbiodinium phylotypes. Most research has been completed on the Symbiodinium clades A–D. Each one of the eight contributes their own benefits as well as less compatible attributes to the survival of their coral hosts. Each photosynthetic organism has a specific level of sensitivity to photodamage of compounds needed for survival, such as proteins. Rates of regeneration and replication determine the organism's ability to survive. Phylotype A is found more in the shallow regions of marine waters. It is able to produce mycosporine-like amino acids that are UV resistant, using a derivative of glycerin to absorb the UV radiation and allowing them to become more receptive to warmer water temperatures. In the event of UV or thermal damage, if and when repair occurs, it will increase the likelihood of survival of the host and symbiont. This leads to the idea that, evolutionarily, clade A is more UV resistant and thermally resistant than the other clades.
Clades B and C are found more frequently in the deeper water regions, which may explain the higher susceptibility to increased temperatures. Terrestrial plants that receive less sunlight because they are found in the undergrowth can be analogized to clades B, C, and D. Since clades B through D are found at deeper depths, they require an elevated light absorption rate to be able to synthesize as much energy. With elevated absorption rates at UV wavelengths, the deeper occurring phylotypes are more prone to coral bleaching versus the more shallow clades. Clade D has been observed to be high temperature-tolerant, and as a result it has a higher rate of survival than clades B and C.
Darwin's paradox
In The Structure and Distribution of Coral Reefs,
published in 1842, Darwin described how coral reefs were found in some
areas of the tropical seas but not others, with no obvious cause. The
largest and strongest corals grew in parts of the reef exposed to the
most violent surf and corals were weakened or absent where loose
sediment accumulated.
Tropical waters contain few nutrients yet a coral reef can flourish like an "oasis in the desert".
This has given rise to the ecosystem conundrum, sometimes called
"Darwin's paradox": "How can such high production flourish in such
nutrient poor conditions?"
Coral reefs cover less than 0.1% of the surface of the world’s ocean,
about half the land area of France, yet they support over one-quarter
of all marine species. This diversity results in complex food webs, with large predator fish eating smaller forage fish that eat yet smaller zooplankton and so on. However, all food webs eventually depend on plants, which are the primary producers. Coral reefs' primary productivity is very high, typically producing 5–10 grams of carbon per square meter per day (gC·m−2·day−1) biomass.
One reason for the unusual clarity of tropical waters is they are deficient in nutrients and drifting plankton. Further, the sun shines year-round in the tropics, warming the surface layer, making it less dense than subsurface layers. The warmer water is separated from deeper, cooler water by a stable thermocline, where the temperature makes a rapid change. This keeps the warm surface waters floating above the cooler deeper waters. In most parts of the ocean, there is little exchange between these layers. Organisms that die in aquatic environments generally sink to the bottom, where they decompose, which releases nutrients in the form of nitrogen (N), phosphorus (P) and potassium (K). These nutrients are necessary for plant growth, but in the tropics, they do not directly return to the surface.
Plants form the base of the food chain, and need sunlight and
nutrients to grow. In the ocean, these plants are mainly microscopic phytoplankton which drift in the water column. They need sunlight for photosynthesis, which powers carbon fixation,
so they are found only relatively near the surface. But they also need
nutrients. Phytoplankton rapidly use nutrients in the surface waters,
and in the tropics, these nutrients are not usually replaced because of
the thermocline.
Explanations
Around
coral reefs, lagoons fill in with material eroded from the reef and the
island. They become havens for marine life, providing protection from
waves and storms.
Most importantly, reefs recycle
nutrients, which happens much less in the open ocean. In coral reefs
and lagoons, producers include phytoplankton, as well as seaweed and
coralline algae, especially small types called turf algae, which pass
nutrients to corals.
The phytoplankton are eaten by fish and crustaceans, who also pass
nutrients along the food web. Recycling ensures fewer nutrients are
needed overall to support the community.
Coral reefs support many symbiotic relationships. In particular, zooxanthellae provide energy to coral in the form of glucose, glycerol, and amino acids. Zooxanthellae can provide up to 90% of a coral’s energy requirements. In return, as an example of mutualism,
the corals shelter the zooxanthellae, averaging one million for every
cubic centimeter of coral, and provide a constant supply of the carbon dioxide they need for photosynthesis.
Corals also absorb nutrients, including inorganic nitrogen and
phosphorus, directly from water. Many corals extend their tentacles at
night to catch zooplankton
that brush them when the water is agitated. Zooplankton provide the
polyp with nitrogen, and the polyp shares some of the nitrogen with the
zooxanthellae, which also require this element.
The varying pigments in different species of zooxanthellae give them an
overall brown or golden-brown appearance, and give brown corals their
colors. Other pigments such as reds, blues, greens, etc. come from
colored proteins made by the coral animals. Coral which loses a large
fraction of its zooxanthellae becomes white (or sometimes pastel shades
in corals that are richly pigmented with their own colorful proteins)
and is said to be bleached, a condition which, unless corrected, can kill the coral.
Sponges are another key: they live in crevices in the coral reefs. They are efficient filter feeders, and in the Red Sea
they consume about 60% of the phytoplankton that drifts by. The sponges
eventually excrete nutrients in a form the corals can use.
The roughness of coral surfaces is the key to coral survival in
agitated waters. Normally, a boundary layer of still water surrounds a
submerged object, which acts as a barrier. Waves breaking on the
extremely rough edges of corals disrupt the boundary layer, allowing the
corals access to passing nutrients. Turbulent water thereby promotes
reef growth and branching. Without the nutritional gains brought by
rough coral surfaces, even the most effective recycling would leave
corals wanting in nutrients.
Studies have shown that deep nutrient-rich water entering coral reefs
through isolated events may have significant effects on temperature and
nutrient systems.
This water movement disrupts the relatively stable thermocline that
usually exists between warm shallow water to deeper colder water.
Leichter et al. (2006)
found that temperature regimes on coral reefs in the Bahamas and
Florida were highly variable with temporal scales of minutes to seasons
and spatial scales across depths.
Water can be moved through coral reefs in various ways, including
current rings, surface waves, internal waves and tidal changes.
Movement is generally created by tides and wind. As tides interact with
varying bathymetry and wind mixes with surface water, internal waves
are created. An internal wave is a gravity wave that moves along density
stratification within the ocean. When a water parcel encounters a
different density it will oscillate and create internal waves.
While internal waves generally have a lower frequency than surface
waves, they often form as a single wave that breaks into multiple waves
as it hits a slope and moves upward. This vertical break up of internal waves causes significant diapycnal mixing and turbulence. Internal waves can act as nutrient pumps, bringing plankton and cool nutrient-rich water up to the surface.
The irregular structure characteristic of coral reef bathymetry may enhance mixing and produce pockets of cooler water and variable nutrient content. Arrival of cool, nutrient-rich water from depths due to internal waves and tidal bores has been linked to growth rates of suspension feeders and benthic algae as well as plankton and larval organisms. Leichter et al. proposed that the seaweed Codium isthmocladum reacts to deep water nutrient sources due to their tissues having different concentrations of nutrients dependent upon depth. Wolanski and Hamner noted aggregations of eggs, larval organisms and plankton on reefs in response to deep water intrusions. Similarly, as internal waves and bores move vertically, surface-dwelling larval organisms are carried toward the shore. This has significant biological importance to cascading effects of food chains in coral reef ecosystems and may provide yet another key to unlocking "Darwin's Paradox".
Cyanobacteria provide soluble nitrates for the reef via nitrogen fixation.
Coral reefs also often depend on surrounding habitats, such as seagrass meadows and mangrove forests,
for nutrients. Seagrass and mangroves supply dead plants and animals
which are rich in nitrogen and also serve to feed fish and animals from
the reef by supplying wood and vegetation. Reefs, in turn, protect
mangroves and seagrass from waves and produce sediment in which the mangroves and seagrass can root.
Biodiversity
Coral reefs form some of the world's most productive ecosystems, providing complex and varied marine habitats that support a wide range of other organisms. Fringing reefs just below low tide level have a mutually beneficial relationship with mangrove forests at high tide level and sea grass meadows in between: the reefs protect the mangroves and seagrass from strong currents and waves that would damage them or erode the sediments in which they are rooted, while the mangroves and sea grass protect the coral from large influxes of silt, fresh water and pollutants. This level of variety in the environment benefits many coral reef animals, which, for example, may feed in the sea grass and use the reefs for protection or breeding.
Reefs are home to a large variety of animals, including fish, seabirds, sponges, cnidarians (which includes some types of corals and jellyfish), worms, crustaceans (including shrimp, cleaner shrimp, spiny lobsters and crabs), mollusks (including cephalopods), echinoderms (including starfish, sea urchins and sea cucumbers), sea squirts, sea turtles and sea snakes. Aside from humans, mammals are rare on coral reefs, with visiting cetaceans such as dolphins being the main exception. A few of these varied species feed directly on corals, while others graze on algae on the reef. Reef biomass is positively related to species diversity.
The same hideouts in a reef may be regularly inhabited by different species at different times of day. Nighttime predators such as cardinalfish and squirrelfish hide during the day, while damselfish, surgeonfish, triggerfish, wrasses and parrotfish hide from eels and sharks.
Algae
Reefs are chronically at risk of algal encroachment. Overfishing and excess nutrient supply from onshore can enable algae to outcompete and kill the coral. Increased nutrient levels can be a result of sewage or chemical fertilizer runoff from nearby coastal developments. Runoff can carry nitrogen and phosphorus which promote excess algae growth. Algae can sometimes out-compete the coral for space. The algae can then smother the coral by decreasing the oxygen supply available to the reef. Decreased oxygen levels can slow down coral's calcification rates weakening the coral and leaving it more susceptible to disease and degradation. In surveys done around largely uninhabited US Pacific islands, algae inhabit a large percentage of surveyed coral locations. The algal population consists of turf algae, coralline algae, and macro algae.Sponges
Sponges are essential for the functioning of the coral reef's ecosystem. Algae and corals in coral reefs produce organic material. This is filtered through sponges which convert this organic material into small particles which in turn are absorbed by algae and corals.Fish
Over 4,000 species of fish inhabit coral reefs. The reasons for this diversity remain unclear. Hypotheses include the "lottery", in which the first (lucky winner) recruit to a territory is typically able to defend it against latecomers, "competition", in which adults compete for territory, and less-competitive species must be able to survive in poorer habitat, and "predation", in which population size is a function of postsettlement piscivore mortality. Healthy reefs can produce up to 35 tons of fish per square kilometer each year, but damaged reefs produce much less.Invertebrates
Sea urchins, Dotidae and sea slugs eat seaweed. Some species of sea urchins, such as Diadema antillarum, can play a pivotal part in preventing algae from overrunning reefs. Nudibranchia and sea anemones eat sponges.A number of invertebrates, collectively called "cryptofauna," inhabit the coral skeletal substrate itself, either boring into the skeletons (through the process of bioerosion) or living in pre-existing voids and crevices. Those animals boring into the rock include sponges, bivalve mollusks, and sipunculans. Those settling on the reef include many other species, particularly crustaceans and polychaete worms.
Seabirds
Coral reef systems provide important habitats for seabird species, some endangered. For example, Midway Atoll in Hawaii supports nearly three million seabirds, including two-thirds (1.5 million) of the global population of Laysan albatross, and one-third of the global population of black-footed albatross. Each seabird species has specific sites on the atoll where they nest. Altogether, 17 species of seabirds live on Midway. The short-tailed albatross is the rarest, with fewer than 2,200 surviving after excessive feather hunting in the late 19th century.Other
Sea snakes feed exclusively on fish and their eggs. Marine birds, such as herons, gannets, pelicans and boobies, feed on reef fish. Some land-based reptiles intermittently associate with reefs, such as monitor lizards, the marine crocodile and semiaquatic snakes, such as Laticauda colubrina. Sea turtles, particularly hawksbill sea turtles, feed on sponges.- The shell of Latiaxis wormaldi, a coral snail
Importance
Coral reefs deliver ecosystem services to tourism, fisheries and coastline protection. The global economic value of coral reefs has been estimated to be between US $29.8 billion and $375 billion per year. Coral reefs protect shorelines by absorbing wave energy, and many small islands would not exist without their reefs to protect them. According to the environmental group World Wide Fund for Nature, the economic cost over a 25-year period of destroying one kilometer of coral reef is somewhere between $137,000 and $1,200,000. About six million tons of fish are taken each year from coral reefs. Well-managed coral reefs have an annual yield of 15 tons of seafood on average per square kilometer. Southeast Asia's coral reef fisheries alone yield about $2.4 billion annually from seafood.To improve the management of coastal coral reefs, another environmental group, the World Resources Institute (WRI) developed and published tools for calculating the value of coral reef-related tourism, shoreline protection and fisheries, partnering with five Caribbean countries. As of April 2011, published working papers covered St. Lucia, Tobago, Belize, and the Dominican Republic, with a paper for Jamaica in preparation. The WRI was also "making sure that the study results support improved coastal policies and management planning". The Belize study estimated the value of reef and mangrove services at $395–559 million annually.
Bermuda's coral reefs provide economic benefits to the Island worth on average $722 million per year, based on six key ecosystem services, according to Sarkis et al (2010).
Threats
Coral reefs are dying around the world. In particular, coral mining, agricultural and urban runoff, pollution (organic and inorganic), overfishing, blast fishing, disease, and the digging of canals
and access into islands and bays are localized threats to coral
ecosystems. Broader threats are sea temperature rise, sea level rise and
pH changes from ocean acidification, all associated with greenhouse gas
emissions. A 2014 study lists factors such as population explosion
along the coast lines, overfishing, the pollution of coastal areas,
global warming and invasive species among the main reasons that have put
reefs in danger of extinction.
A study released in April 2013 has shown that air pollution can also
stunt the growth of coral reefs; researchers from Australia, Panama and
the UK used coral records (between 1880 and 2000) from the western
Caribbean to show the threat of factors such as coal-burning coal and
volcanic eruptions. Pollutants, such as Tributyltin, a biocide released into water from in anti-fouling paint can be toxic to corals.
In 2011, researchers suggested that "extant marine invertebrates face
the same synergistic effects of multiple stressors" that occurred
during the end-Permian extinction,
and that genera "with poorly buffered respiratory physiology and
calcareous shells", such as corals, were particularly vulnerable.
Rock coral on seamounts across the ocean are under fire from bottom trawling.
Reportedly up to 50% of the catch is rock coral, and the practice
transforms coral structures to rubble. With it taking years to regrow,
these coral communities are disappearing faster than they can sustain
themselves.
Another cause for the death of coral reefs is bioerosion.
Various fishes graze corals, dead or alive and change the morphology of
coral reefs making them more susceptible to other physical and chemical
threats. It has been generally observed that only the algae growing on
dead corals is eaten and the live ones are not. However, this act still
destroys the top layer of coral substrate and makes it harder for the
reefs to sustain.
In El Niño-year 2010, preliminary reports show global coral bleaching
reached its worst level since another El Niño year, 1998, when 16% of
the world's reefs died as a result of increased water temperature. In
Indonesia's Aceh
province, surveys showed some 80% of bleached corals died. Scientists
do not yet understand the long-term impacts of coral bleaching, but they
do know that bleaching leaves corals vulnerable to disease, stunts
their growth, and affects their reproduction, while severe bleaching
kills them. In July, Malaysia closed several dive sites where virtually all the corals were damaged by bleaching.
To find answers for these problems, researchers study the various
factors that impact reefs. The list includes the ocean's role as a carbon dioxide sink, atmospheric changes, ultraviolet light, ocean acidification, viruses, impacts of dust storms carrying agents to far-flung reefs, pollutants, algal blooms and others. Reefs are threatened well beyond coastal areas. Coral reefs with one type of zooxanthellae are more prone to bleaching than are reefs with another, more hardy, species.
General estimates show approximately 10% of the world's coral reefs are dead.
About 60% of the world's reefs are at risk due to destructive,
human-related activities. The threat to the health of reefs is
particularly high in Southeast Asia, where 95% of reefs are at risk from local threats. By the 2030s, 90% of reefs are expected to be at risk from both human activities and climate change; by 2050, all coral reefs will be in danger.
Current research is showing that ecotourism in the Great Barrier Reef is contributing to coral disease, and that chemicals in sunscreens may contribute to the impact of viruses on zooxanthellae.
Some scientists, including those associated with the National Oceanic and Atmospheric Administration, posit that US coral reefs are likely to disappear within a few decades as a result of global warming.
Protection

Marine protected areas (MPAs) have become increasingly prominent for reef management. MPAs promote responsible fishery management and habitat protection. Much like national parks and wildlife refuges, and to varying degrees, MPAs restrict potentially damaging activities. MPAs encompass both social and biological objectives, including reef restoration, aesthetics, biodiversity, and economic benefits. However, there are very few MPAs that have actually made a substantial difference. Research in Indonesia, Philippines and Papua New Guinea shows that there is no significant difference between an MPA site and an unprotected site. Conflicts surrounding MPAs involve lack of participation, clashing views of the government and fisheries, effectiveness of the area, and funding. In some situations, as in the Phoenix Islands Protected Area, MPAs can also provide revenue, potentially equal to the income they would have generated without controls, as Kiribati did for its Phoenix Islands.
According to the Caribbean Coral Reefs - Status Report 1970-2012 made by the IUCN. States that; stopping overfishing especially key fishes to coral reef like parrotfish, coastal zone management which reduce human pressure on reef, (for example restricting the coastal settlement, development and tourism in coastal reef) and controlling pollution specially sewage wastage, may not only reduce coral declining but also reverse it and may let to coral reef more adaptable to changes relates to climate and acidification. The report shows that healthier reef in the Caribbean are those with large population of parrotfish in countries which protect these key fishes and sea urchins, banning fish trap and Spearfishing creating "resilient reefs".
To help combat ocean acidification, some laws are in place to reduce greenhouse gases such as carbon dioxide. The Clean Water Act puts pressure on state government agencies to monitor and limit runoff of pollutants that can cause ocean acidification. Stormwater surge preventions are also in place, as well as coastal buffers between agricultural land and the coastline. This act also ensures that delicate watershed ecosystems are intact, such as wetlands. The Clean Water Act is funded by the federal government, and is monitored by various watershed groups. Many land use laws aim to reduce CO2 emissions by limiting deforestation. Deforestation causes erosion, which releases a large amount of carbon stored in the soil, which then flows into the ocean, contributing to ocean acidification. Incentives are used to reduce miles traveled by vehicles, which reduces the carbon emissions into the atmosphere, thereby reducing the amount of dissolved CO2 in the ocean. State and federal governments also control coastal erosion, which releases stored carbon in the soil into the ocean, increasing ocean acidification. High-end satellite technology is increasingly being employed to monitor coral reef conditions.
Biosphere reserve, marine park, national monument and world heritage status can protect reefs. For example, Belize's barrier reef, Sian Ka'an, the Galapagos islands, Great Barrier Reef, Henderson Island, Palau and Papahānaumokuākea Marine National Monument are world heritage sites.
In Australia, the Great Barrier Reef is protected by the Great Barrier Reef Marine Park Authority, and is the subject of much legislation, including a biodiversity action plan. They have compiled a Coral Reef Resilience Action Plan. This detailed action plan consists of numerous adaptive management strategies, including reducing our carbon footprint, which would ultimately reduce the amount of ocean acidification in the oceans surrounding the Great Barrier Reef. An extensive public awareness plan is also in place to provide education on the “rainforests of the sea” and how people can reduce carbon emissions, thereby reducing ocean acidification.
Inhabitants of Ahus Island, Manus Province, Papua New Guinea, have followed a generations-old practice of restricting fishing in six areas of their reef lagoon. Their cultural traditions allow line fishing, but no net or spear fishing. The result is both the biomass and individual fish sizes are significantly larger than in places where fishing is unrestricted.
Restoration
Coral aquaculture, also known as coral farming or coral gardening, is showing promise as a potentially effective tool for restoring coral reefs, which have been declining around the world. The process bypasses the early growth stages of corals when they are most at risk of dying. Coral seeds are grown in nurseries, then replanted on the reef. Coral is farmed by coral farmers who live locally to the reefs and farm for reef conservation or for income.
Efforts to expand the size and number of coral reefs generally involve supplying substrate to allow more corals to find a home. Substrate materials include discarded vehicle tires, scuttled ships, subway cars, and formed concrete, such as reef balls. Reefs also grow unaided on marine structures such as oil rigs. In large restoration projects, propagated hermatypic coral on substrate can be secured with metal pins, superglue or milliput. Needle and thread can also attach A-hermatype coral to substrate.
A substrate for growing corals referred to as Biorock is produced by running low voltage electrical currents through seawater to crystallize dissolved minerals onto steel structures. The resultant white carbonate (aragonite) is the same mineral that makes up natural coral reefs. Corals rapidly colonize and grow at accelerated rates on these coated structures. The electrical currents also accelerate formation and growth of both chemical limestone rock and the skeletons of corals and other shell-bearing organisms. The vicinity of the anode and cathode provides a high-pH environment which inhibits the growth of competitive filamentous and fleshy algae. The increased growth rates fully depend on the accretion activity.
During accretion, the settled corals display an increased growth rate, size and density, but after the process is complete, growth rate and density return to levels comparable to natural growth, and are about the same size or slightly smaller.
One case study with coral reef restoration was conducted on the island of Oahu in Hawaii. The University of Hawaii has come up with a Coral Reef Assessment and Monitoring Program to help relocate and restore coral reefs in Hawaii. A boat channel on the island of Oahu to the Hawaii Institute of Marine Biology was overcrowded with coral reefs. Also, many areas of coral reef patches in the channel had been damaged from past dredging in the channel. Dredging covers the existing corals with sand, and their larvae cannot build and thrive on sand; they can only build on to existing reefs. Because of this, the University of Hawaii decided to relocate some of the coral reef to a different transplant site. They transplanted them with the help of the United States Army divers, to a relocation site relatively close to the channel. They observed very little, if any, damage occurred to any of the colonies while they were being transported, and no mortality of coral reefs has been observed on the new transplant site, but they will be continuing to monitor the new transplant site to see how potential environmental impacts (i.e. ocean acidification) will harm the overall reef mortality rate. While trying to attach the coral to the new transplant site, they found the coral placed on hard rock is growing considerably well, and coral was even growing on the wires that attached the transplant corals to the transplant site. This gives new hope to future research on coral reef transplant sites. As a result of this coral restoration project, no environmental effects were seen from the transplantation process, no recreational activities were decreased, and no scenic areas were affected by the project. This is a great example that coral transplantation and restoration can work and thrive under the right conditions, which means there may be hope for other damaged coral reefs.
Another possibility for coral restoration is gene therapy. Through infecting coral with genetically modified bacteria, it may be possible to grow corals that are more resistant to climate change and other threats.
Reefs in the past
Throughout Earth history, from a few thousand years after hard skeletons were developed by marine organisms, there were almost always reefs. The times of maximum development were in the Middle Cambrian (513–501 Ma), Devonian (416–359 Ma) and Carboniferous (359–299 Ma), owing to order Rugosa extinct corals, and Late Cretaceous (100–66 Ma) and all Neogene (23 Ma–present), owing to order Scleractinia corals.
Not all reefs in the past were formed by corals: those in the Early Cambrian (542–513 Ma) resulted from calcareous algae and archaeocyathids (small animals with conical shape, probably related to sponges) and in the Late Cretaceous (100–66 Ma), when there also existed reefs formed by a group of bivalves called rudists; one of the valves formed the main conical structure and the other, much smaller valve acted as a cap.
Measurements of the oxygen isotopic composition of the aragonitic skeleton of coral reefs, such as Porites, can indicate changes in the sea surface temperature and sea surface salinity conditions of the ocean during the growth of the coral. This technique is often used by climate scientists to infer the paleoclimate of a region.