The taxonomic classification of humans following John Edward Gray (1825).
Overview of speciation and hybridization within the genus Homo over the last two million years (vertical axis). The rapid "Out of Africa" expansion of H. sapiens is indicated at the top of the diagram, with admixture indicated with Neanderthals, Denisovans, and unspecified archaic African hominins.
| Homo ("humans") | |
|---|---|
| Scientific classification | |
| Kingdom: | Animalia |
| Phylum: | Chordata |
| Class: | Mammalia |
| Order: | Primates |
| Suborder: | Haplorhini |
| Infraorder: | Simiiformes |
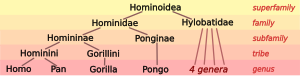
| Family: | Hominidae |
| Subfamily: | Homininae |
| Tribe: | Hominini |
| Genus: | Homo Linnaeus, 1758 |
| Type species | |
| Homo sapiens
Linnaeus, 1758
| |
| Species | |
|
Homo sapiens †Homo erectus other species or subspecies suggested | |
Human taxonomy is the classification of the human species (systematic name Homo sapiens) within zoological taxonomy. The systematic genus, Homo, is designed to include both anatomically modern humans and extinct varieties of archaic humans.
Since the introduction of systematic names in the 18th century, knowledge of human evolution has increased drastically, and a number of intermediate taxa have been proposed in the 20th to early 21st century. The most widely accepted taxonomy groups takes the genus Homo as originating between two and three million years ago, divided into at least two species, archaic Homo erectus and modern Homo sapiens, with about a dozen further suggestions for species without universal recognition.
The genus Homo is placed in the tribe Hominini alongside Pan (chimpanzees). The two genera are estimated to have diverged over an extended time of hybridization spanning roughly 10 to 6 million years ago, with possible admixture as late as 4 million years ago. A subtribe of uncertain validity, grouping archaic "pre-human" or "para-human" species younger than the Homo-Pan split is Australopithecina (proposed in 1939).
A proposal by Wood and Richmond (2000) would introduce Hominina as a subtribe alongside Australopithecina, with Homo the only known genus within Hominina. Alternatively, following Cela-Conde and Ayala (2003), the "pre-human" or "proto-human" genera of Australopithecus, Ardipithecus, Praeanthropus, and possibly Sahelanthropus may be placed on equal footing alongside the genus Homo. An even more radical view rejects the division of Pan and Homo as separate genera, which based on the Principle of Priority would imply the re-classification of chimpanzees as Homo paniscus (or similar).
History
Human taxonomy on one hand involves the placement of humans within the Taxonomy of the hominids (great apes), and on the other the division of archaic and modern humans into species and, if applicable, subspecies. Modern zoological taxonomy was developed by Carl Linnaeus during the 1730s to 1750s. He named the human species as Homo sapiens in 1758, as the only member species of the genus Homo, divided into several subspecies corresponding to the great races. The Latin noun homō (genitive hominis) means "human being". The systematic name Hominidae for the family of the great apes was introduced by John Edward Gray (1825). Gray also supplied Hominini as the name of the tribe including both chimpanzees (genus Pan) and humans (genus Homo).The discovery of the first extinct archaic human species from the fossil record dates to the mid 19th century, Homo neanderthalensis, classified in 1864. Since then, a number of other archaic species have been named, but there is no universal consensus as to their exact number. After the discovery of H. neanderthalensis, which even if "archaic" is recognizable as clearly human, late 19th to early 20th century anthropology for a time was occupied with finding the supposedly "missing link" between Homo and Pan. The "Piltdown Man" hoax of 1912 was the (fraudulent) presentation of such a transitional species. Since the mid-20th century, knowledge of the development of Hominini has become much more detailed, and taxonomical terminology has been altered a number of times to reflect this.
The introduction of Australopithecus as a third genus, alongside Homo and Pan, in the Hominini tribe is due to Raymond Dart (1925). Australopithecina as a subtribe containing Australopithecus as well as Paranthropus (Broom 1938) is a proposal by Gregory & Hellman (1939). More recently proposed additions to the Australopithecina subtribe include Ardipithecus (1995) and Kenyanthropus (2001). The position of Sahelanthropus (2002) relative to Australopithecina within Hominini is unclear. Cela-Conde and Ayala (2003) propose the recognition of Australopithecus, Ardipithecus, Praeanthropus, and Sahelanthropus (the latter incertae sedis) as separate genera.
Other proposed genera, now mostly considered part of Homo, include: Pithecanthropus (Dubois, 1894), Protanthropus (Haeckel, 1895), Sinanthropus (Black, 1927), Cyphanthropus (Pycraft, 1928) Africanthropus (Dreyer, 1935), Telanthropus (Broom & Anderson 1949), Atlanthropus (Arambourg, 1954), Tchadanthropus (Coppens, 1965).
The genus Homo has been taken to originate some two millions ago since the discovery of stone tools in Olduvai Gorge, Tanzania, in the 1960s. Homo habilis (Leakey et al., 1964) would be the first "human" species (member of genus Homo) by definition, its type specimen being the OH 7 fossils. However, the discovery of more fossils of this type has opened up the debate on the delineation of H. habilis from Australopithecus. Especially, the LD 350-1 jawbone fossil discovered in 2013, dated to 2.8 Mya, has been argued as being transitional between the two. It is also disputed whether H. habilis was the first hominin to use stone tools, as Australopithecus garhi, dated to c. 2.5 Mya, has been found along with stone tool implements. Fossil KNM-ER 1470 (discovered in 1972, designated Pithecanthropus rudolfensis by Alekseyev 1978) is now seen as either a third early species of Homo (alongside H. habilis and H. erectus) at about 2 million years ago, or alternatively as transitional between Australopithecus and Homo.
Wood and Richmond (2000) proposed that Gray's tribe Hominini ("hominins") be designated as comprising all species after the chimpanzee-human last common ancestor by definition, to the inclusion of Australopithecines and other possible pre-human or para-human species (such as Ardipithecus and Sahelanthropus) not known in Gray's time. In this suggestion, the new subtribe of Hominina was to be designated as including the genus Homo exclusively, so that Hominini would have two subtribes, Australopithecina and Hominina, with the only known genus in Hominina being Homo. Orrorin (2001) has been proposed as a possible ancestor of Hominina but not Australopithecina.
Designations alternative to Hominina have been proposed: Australopithecinae (Gregory & Hellman 1939) and Preanthropinae (Cela-Conde & Altaba 2002);
Species
At least a dozen species of Homo other than Homo sapiens have been proposed, with varying degrees of consensus. Homo erectus is widely recognized as the species directly ancestral to Homo sapiens. Most other proposed species are proposed as alternatively belonging to either Homo erectus or Homo sapiens as a subspecies. This concerns Homo ergaster in particular. One proposal divides Homo erectus into an African and an Asian variety; the African is Homo ergaster, and the Asian is Homo erectus sensu stricto. (Inclusion of Homo ergaster with Asian Homo erectus is Homo erectus sensu lato.) There appears to be a recent trend, with the availability of ever more difficult-to-classify fossils such as the Dmanisi skulls (2013) or Homo naledi fossils (2015) to subsume all archaic varieties under Homo erectus.Subspecies
Homo sapiens subspecies
The recognition or non-recognition of subspecies of Homo sapiens has a complicated history. The rank of subspecies in zoology is introduced for convenience, and not by objective criteria, based on pragmatic consideration of factors such as geographic isolation and sexual selection. The informal taxonomic rank of race is variously considered equivalent or subordinate to the rank of subspecies, and the division of anatomically modern humans (H. sapiens) into subspecies is closely tied to the recognition of major racial groupings based on human genetic variation.
A subspecies cannot be recognized independently: a species will either be recognized as having no subspecies at all or at least two (including any that are extinct). Therefore, the designation of an extant subspecies Homo sapiens sapiens only makes sense if at least one other subspecies is recognized. H. s. sapiens is attributed to "Linnaeus (1758)" by the taxonomic Principle of Coordination. William Stearn (1959) in a "passing remark" argued that Linnaeus "must stand as the type of his Homo sapiens". Since Linnaeus describes H. s. europaeus as having blue/green (caerulus) eyes but himself had brown eyes, he cannot have included himself in H. s. europaeus, Linnaeus would therefore have to be classified as H. sapiens sapiens, as not matching any of the descriptions of his five subspecies, and so would stand as the lectotype both for H. sapiens, and for H. s. sapiens within his own subspecies nomenclature.
During the 19th to mid-20th century, it was common practice to classify the major divisions of extant H. sapiens as subspecies, following Linnaeus (1758), who had recognized H. s. americanus, H. s. europaeus, H. s. asiaticus and H. s. afer as grouping the native populations of the Americas, West Eurasia, East Asia and Sub-Saharan Africa, respectively, besides H. s. ferus (for the "wild" form which he identified with feral children) and two further "wild" forms for reported specimens now considered part of cryptozoology, H. s. monstrosus and H. s. troglodytes.
There were variations and additions to the categories of Linnaeus, such as H. s. tasmanianus for the native population of Australia. Bory de St. Vincent in his Essai sur l'Homme (1825) extended Linné's "racial" categories to as many as fifteen: Leiotrichi ("smooth-haired"): japeticus (with subraces), arabicus, indicus, scythicus, sinicus, hyperboreus, neptunianus, australasicus, columbicus, americanus, patagonicus; Oulotrichi ("crisp-haired"): aethiopicus, cafer, hottentotus, melaninus. Similarly, Georges Vacher de Lapouge (1899) also had categories based on race, such as priscus, spelaeus (etc.).
Homo sapiens neanderthalensis was proposed by King (1864) as an alternative to Homo neanderthalensis. There have been "taxonomic wars" over whether Neanderthals were a separate species since their discovery in the 1860s. Pääbo (2014) frames this as a debate that is unresolvable in principle, "since there is no definition of species perfectly describing the case." Louis Lartet (1869) proposed Homo sapiens fossilis based on the Cro-Magnon fossils.
There are a number of proposals of extinct varieties of Homo sapiens made in the 20th century. Many of the original proposals were not using explicit trinomial nomenclature, even though they are still cited as valid synonyms of H. sapiens by Wilson & Reeder (2005). These include: Homo grimaldii (Lapouge, 1906), Homo aurignacensis hauseri (Klaatsch & Hauser, 1910), Notanthropus eurafricanus (Sergi, 1911), Homo fossilis infrasp. proto-aethiopicus (Giuffrida-Ruggeri, 1915), Telanthropus capensis (Broom, 1917),[39] Homo wadjakensis (Dubois, 1921), Homo sapiens cro-magnonensis, Homo sapiens grimaldiensis (Gregory, 1921), Homo drennani (Kleinschmidt, 1931), Homo galilensis (Joleaud, 1931) = Paleanthropus palestinus (McCown & Keith, 1932). Rightmire (1983) proposed Homo sapiens rhodesiensis.
By the 1980s, the practice of dividing extant populations of Homo sapiens into subspecies declined. An early authority explicitly avoiding the division of H. sapiens into subspecies was Grzimeks Tierleben, published 1967–1972. A late example of an academic authority proposing that the human racial groups should be considered taxonomical subspecies is John Baker (1974). The trinomial nomenclature Homo sapiens sapiens became popular for "modern humans" in the context of Neanderthals being considered a subspecies of H. sapiens in the second half of the 20th century. Derived from the convention, widespread in the 1980s, of considering two subspecies, H. s. neanderthalensis and H. s. sapiens, the explicit claim that "H. s. sapiens is the only extant human subspecies" appears in the early 1990s. This is only true if the nomenclature derived from Linnaeus is rejected. Based on Linnaeus (1758), there are at least six subspecies, with H. s. sapiens catching those specimens not included in any other.
Since the 2000s, the extinct Homo sapiens idaltu (White et al., 2003) has gained wide recognition as a subspecies of Homo sapiens, but even in this case there is a dissenting view arguing that "the skulls may not be distinctive enough to warrant a new subspecies name". H. s. neanderthalensis and H. s. rhodesiensis continue to be considered separate species by some authorities, but the genetic evidence of archaic human admixture with modern humans discovered in the 2010s has re-opened the details of taxonomy of archaic humans.
Homo erectus ssp.
Homo erectus since its introduction in 1892 has been divided into numerous subspecies, many of them formerly considered individual species of Homo. None of these subspecies have universal consensus among paleontologists.- Homo erectus erectus (Java Man) (1970s)
- Homo erectus yuanmouensis (Yuanmou Man) (Li et al., 1977)
- Homo erectus lantianensis (Lantian Man) (Woo Ju-Kang, 1964)
- Homo erectus nankinensis (Nanjing Man) (1993)
- Homo erectus pekinensis (Peking Man) (1970s)
- Homo erectus palaeojavanicus (Meganthropus) (Tyler, 2001)
- Homo erectus soloensis (Solo Man) (Oppenoorth, 1932)
- Homo erectus tautavelensis (Tautavel Man) (de Lumley and de Lumley, 1971)
- Homo erectus georgicus (1991)
- Homo erectus bilzingslebenensis (Vlček, 2002)