A genealogical DNA test is a DNA-based test which looks at specific locations of a person's genome, in order to find or verify ancestral genealogical relationships or (with lower reliability) to estimate the ethnic
mixture of an individual. Since different testing companies use
different ethnic reference groups and different matching algorithms,
ethnicity estimates for an individual will vary between tests, sometimes
dramatically.
Three principal types of genealogical DNA tests are available, with each looking at a different part of the genome and useful for different types of genealogical research: autosomal, mitochondrial (mtDNA), and Y-DNA.
Autosomal tests may result in a large amount of DNA matches (other test persons that the individual may be related to), along mixed male and female lines, each match with an estimated distance in the family tree. However, due to the random nature of which and how much DNA is inherited by each tested person from their common ancestors, precise conclusions can only be made for close relations. Traditional genealogical research, and the sharing of family trees, is typically required for interpretation of the results. Autosomal tests are also used in estimating ethnic mix.
MtDNA and Y-DNA tests are much more objective. However, they give considerably fewer DNA matches, if any, since they are limited to relationships along a strict female line and a strict male line respectively. MtDNA and Y-DNA tests are utilized to identify archeological cultures and migration paths of a person's ancestors along a strict mother's line or a strict father's line. Based on MtDNA and Y-DNA, a person's haplogroup(s) can be identified. Only men can take Y-DNA tests, since women lack a Y chromosome.
Three principal types of genealogical DNA tests are available, with each looking at a different part of the genome and useful for different types of genealogical research: autosomal, mitochondrial (mtDNA), and Y-DNA.
Autosomal tests may result in a large amount of DNA matches (other test persons that the individual may be related to), along mixed male and female lines, each match with an estimated distance in the family tree. However, due to the random nature of which and how much DNA is inherited by each tested person from their common ancestors, precise conclusions can only be made for close relations. Traditional genealogical research, and the sharing of family trees, is typically required for interpretation of the results. Autosomal tests are also used in estimating ethnic mix.
MtDNA and Y-DNA tests are much more objective. However, they give considerably fewer DNA matches, if any, since they are limited to relationships along a strict female line and a strict male line respectively. MtDNA and Y-DNA tests are utilized to identify archeological cultures and migration paths of a person's ancestors along a strict mother's line or a strict father's line. Based on MtDNA and Y-DNA, a person's haplogroup(s) can be identified. Only men can take Y-DNA tests, since women lack a Y chromosome.
DNA testing for consumers
The first company to provide direct-to-consumer genetic DNA testing was the now defunct GeneTree. However, it did not offer multi-generational genealogy tests. In fall 2001, GeneTree sold its assets to Salt Lake City-based Sorenson Molecular Genealogy Foundation (SMGF) which originated in 1999.
While in operation, SMGF provided free Y-Chromosome and mitochondrial DNA tests to thousands.
Later, GeneTree returned to genetic testing for genealogy in conjunction
with the Sorenson parent company and eventually was part of the assets
acquired in the Ancestry.com buyout of SMGF in 2012.
In 2000, Family Tree DNA, founded by Bennett Greenspan
and Max Blankfeld, was the first company dedicated to
direct-to-consumer testing for genealogy research. They initially
offered eleven marker Y-Chromosome STR tests and HVR1 mitochondrial DNA
tests. They originally tested in partnership with the University of
Arizona.
In 2007, 23andMe was the first company to offer a saliva-based direct-to-consumer genetic testing. It was also the first to implement using autosomal DNA for ancestry testing, which all other major companies now use.
MyHeritage launched its genetic testing service in 2016, allowing users to use cheek swabs to collect samples. In 2019, new analysis tools were presented: autoclusters (grouping all matches visually into clusters)
and family tree theories (suggesting conceivable relations between DNA
matches by combining several Myheritage trees as well as the Geni global
family tree).
Living DNA, founded in 2015, also provides a genetic testing
service. Living DNA uses SNP chips to provide reports on autosomal
ancestry, Y, and mtDNA ancestry. Living DNA provides detailed reports on ancestry from the UK as well as detailed Y chromosome and mtDNA reports.
In 2019 it was estimated that large genealogical testing companies had about 26 million DNA profiles. Many transferred their test result for free to multiple testing sites, and also to genealogical services such as Geni.com and GEDmatch. GEDMatch said half of their profiles were from the USA.
The popular consciousness of DNA testing and of DNA generally is
subject to a number of misconceptions involving the reliability of
testing, the nature of the connections with one's ancestors, the
connection between DNA and personal traits, etc.
Procedure
A hospital corpsman uses a swab to take a DNA sample from a sailor aboard USS Iwo Jima (LHD 7)
A genealogical DNA test is performed on a DNA sample. This DNA sample can be obtained by a cheek-scraping (also known as a buccal swab), spit-cups, mouthwash, and chewing gum. Typically, the sample collection uses a home test kit supplied by a service provider such as 23andMe, AncestryDNA, Family Tree DNA, or MyHeritage. After following the kit instructions on how to collect the sample, it is returned to the supplier for analysis.
Types of tests
There are three major types of genealogical DNA tests: Autosomal and X-DNA, Y-DNA and mtDNA.
- Autosomal tests look at chromosomes 1–22 and X. The autosomes (chromosomes 1–22) are inherited from both parents and all recent ancestors. The X-chromosome follows a special inheritance pattern. Ethnicity estimates are often included with this sort of testing.
- Y-DNA looks at the Y-chromosome, which is inherited father to son, and so can only be taken by males to explore their direct paternal line.
- mtDNA looks at the mitochondria, which is inherited from mother to child and so can be used to explore one's direct maternal line.
Y-DNA and mtDNA cannot be used for ethnicity estimates, but can be used to find one's haplogroup, which is unevenly distributed geographically.
Direct-to-consumer DNA test companies have often labeled haplogroups by
continent or ethnicity (e.g., an "African haplogroup" or a "Viking
haplogroup"), but these labels may be speculative or misleading.
Autosomal DNA (atDNA) testing
Testing
Autosomal DNA is contained in the 22 pairs of chromosomes not involved in determining a person's sex. Autosomal DNA recombines each generation, and new offspring receive one set of chromosomes from each parent. These are inherited exactly equally from both parents and roughly equally from grandparents to about 3x great-grand parents. Therefore, the number of markers (one of two or more known variants in the genome at a particular location – known as Single-nucleotide polymorphisms
or SNPs) inherited from a specific ancestor decreases by about half
each generation; that is, an individual receives half of their markers
from each parent, about a quarter of their markers from each
grandparent; about an eighth of their markers from each great
grandparent, etc. Inheritance is more random and unequal from more
distant ancestors. Generally, a genealogical DNA test might test about 700,000 SNPs (specific points in the genome).
Shared DNA for different relatives
Reporting process
The preparation of a report on the DNA in the sample proceeds in multiple stages:
- identification of the DNA base pair at specific SNP locations
- comparison with previously stored results
- interpretation of matches
Base pair identification
All major service providers use equipment with chips supplied by Illumina.
The chip determines which SNP locations are tested. Different versions
of the chip are used by different service providers. In addition,
updated versions of the Illumina chip may test different sets of SNP
locations. The list of SNP locations and base pairs at that location is
usually available to the customer as "raw data". The raw data can
sometimes be uploaded to another service provider to produce an
additional interpretation and matches. For additional analysis the data
can also be uploaded to GEDmatch (a third-party web based set of tools
that analyzes raw data from the main service providers).
Identification of Matches
The
major component of an autosomal DNA test is matching other individuals.
Where the individual being tested has a number of consecutive SNPs in
common with a previously tested individual in the company's database, it
can be inferred that they share a segment of DNA at that part of their
genomes.
If the segment is longer than a threshold amount set by the testing
company, then these two individuals are considered to be a match. Unlike
the identification of base pairs, the data bases against which the new
sample is tested, and the algorithms used to determine a match, are
proprietary and specific to each company.
The unit for segments of DNA is the centimorgan
(cM). For comparison, a full human genome is about 6500 cM. The shorter
the length of a match, the greater are the chances that a match is
spurious.
An important statistic for subsequent interpretation is the length of
the shared DNA (or the percentage of the genome that is shared).
Interpretation of Autosomal matches
Most
companies will show the customers how many cMs they share, and across
how many segments. From the number of cMs and segments, the relationship
between the two individuals can be estimated, however due to the random
nature of DNA inheritance, relationship estimates, especially for
distant relatives, are only approximate. Some more distant cousins will
not match at all.
Although information about specific SNPs can be used for some purposes
(eg suggesting likely eye colour), the key information is the percentage
of DNA shared by 2 individuals. This can indicate the closeness of the
relationship. However, it does not show the roles of the 2 individuals -
eg 50% shared suggests a parent - child relationship, but does not
identify which individual is the parent.
Various advanced techniques and analysis can be done on this data. This includes features such as In-common/Shared Matches, Chromosome Browsers and Triangulation. This analysis is often required if DNA evidence is being used to prove or disprove a specific relationship.
X-chromosome DNA testing
The
X-chromosome SNP results are often included in Autosomal DNA tests.
Both males and females receive an X-chromosome from their mother, but
only females receive a second X-chromosome from their father.
The X-chromosome has a special path of inheritance patterns and can be
useful in significantly narrowing down possible ancestor lines
compared to Autosomal DNA – for example an X-chromosome match with a
male can only have come from his maternal side.
Like autosomal DNA, X-chromosome DNA undergoes random recombination at
each generation (except for father to daughter X-chromosomes which are
passed down unchanged). There are specialised inheritance charts which
describe the possible patterns of X-chromosome DNA inheritance for males
and females.
STRs
Some genealogical companies offered autosomal STRs (short tandem repeats). These are similar to Y-DNA STRs. The number of STRs offered is limited, and results have been used for personal identification, paternity cases and inter-population studies.
Law enforcement agencies in the US and Europe use autosomal STR data to identify criminals.
Mitochondrial DNA (mtDNA) testing
The mitochondrion
is a component of a human cell, and contains its own DNA. Mitochondrial
DNA usually has 16,569 base pairs (the number can vary slightly
depending on addition or deletion mutations)
and is much smaller than the human genome DNA which has 3.2 billion
base pairs. Mitochondrial DNA is transmitted from mother to child, thus
a direct maternal ancestor can be traced using mtDNA.
The transmission occurs with relatively rare mutations compared to the
genome DNA. A perfect match found to another person's mtDNA test results
indicates shared ancestry of possibly between 1 and 50 generations ago. More distant matching to a specific haplogroup or subclade may be linked to a common geographic origin.
Test
mtDNA, by current conventions, is divided into three regions. They are the coding region (00577-16023) and two Hyper Variable Regions (HVR1 [16024-16569], and HVR2 [00001-00576]).
The two most common mtDNA tests are a sequence of HVR1 and HVR2
and a full sequence of the mitochondria. Generally, testing only the
HVRs has limited genealogical use so it is increasingly popular and
accessible to have a full sequence. The full mtDNA sequence is only
offered by Family Tree DNA among the major testing companies and is somewhat controversial because the coding region DNA may reveal medical information about the test-taker.
Haplogroups
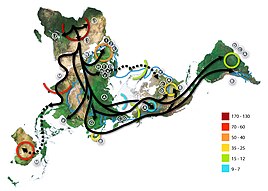
Map of human migration out of Africa,
according to Mitochondrial DNA. The numbers represent thousands of
years before present time. The blue line represents the area covered in
ice or tundra during the last great ice age. The North Pole is at the
center. Africa, the center of the start of the migration, is at the top
left and South America is at the far right.
All humans descend in the direct female line from Mitochondrial Eve,
a female who lived probably around 200,000 years ago in Africa.
Different branches of her descendants are different haplogroups. Most
mtDNA results include a prediction or exact assertion of one's mtDNA Haplogroup. Mitochrondial haplogroups were greatly popularized by the book The Seven Daughters of Eve, which explores mitochondrial DNA.
Understanding mtDNA test results
It is not normal for test results to give a base-by-base list of results. Instead, results are normally compared to the Cambridge Reference Sequence
(CRS), which is the mitochondria of a European who was the first person
to have their mtDNA published in 1981 (and revised in 1999).
Differences between the CRS and testers are usually very few, thus it
is more convenient than listing one's raw results for each base pair.
- Examples
Note that in HVR1, instead of reporting the base pair exactly, for
example 16,111, the 16 is often removed to give in this example 111. The
letters refer to one of the four bases (A, T, G, C) that make up DNA.
| Region | HVR1 | HVR2 |
|---|---|---|
| Differences from CRS | 111T,223T,259T,290T,319A,362C | 073G,146C,153G |
Y chromosome (Y-DNA) testing
The
Y-Chromosome is one of the 23rd pair of human chromosomes. Only males
have a Y-chromosome, because women have two X chromosomes in their 23rd
pair. A man's patrilineal ancestry, or male-line ancestry, can be traced using the DNA on his Y chromosome (Y-DNA), because the Y-chromosome is transmitted father to son nearly unchanged. A man's test results are compared to another man's results to determine the time frame in which the two individuals shared a most recent common ancestor,
or MRCA, in their direct patrilineal lines. If their test results are
very close, they are related within a genealogically useful time frame. A surname project is where many individuals whose Y-chromosomes match collaborate to find their common ancestry.
Women who wish to determine their direct paternal DNA ancestry
can ask their father, brother, paternal uncle, paternal grandfather, or a
paternal uncle's son (their cousin) to take a test for them.
There are two types of DNA testing: STRs and SNPs.
STR markers
Most common is STRs
(short tandem repeat). A certain section of DNA is examined for a
pattern that repeats (e.g. ATCG). The number of times it repeats is the
value of the marker. Typical tests test between 12 and 111 STR markers.
STRs mutate fairly frequently. The results of two individuals are then
compared to see if there is a match. DNA companies will usually provide
an estimate of how closely related two people are, in terms of
generations or years, based on the difference between their results.
SNP markers and Haplogroups
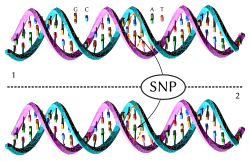
Strand 1 differs from strand 2 at a single base pair location (a C → T polymorphism).
A person's haplogroup can often be inferred from their STR results, but can be proven only with a Y-chromosome SNP test (Y-SNP test).
A single-nucleotide polymorphism (SNP) is a change to a single nucleotide in a DNA sequence. Typical Y-DNA SNP tests test about 20,000 to 35,000 SNPs.
Getting a SNP test allows a much higher resolution than STRs. It can be
used to provide additional information about the relationship between
two individuals and to confirm haplogroups.
The most common Y-DNA-haplogroup in different regions in Europe
All human men descend in the paternal line from a single man dubbed Y-chromosomal Adam,
who lived probably between 200,000 and 400,000 years ago. A 'family
tree' can be drawn showing how men today descend from him. Different
branches of this tree are different haplogroups. Most haplogroups can be
further subdivided multiple times into sub-clades. Some known
sub-clades were founded in the last 1000 years, meaning their timeframe
approaches the genealogical era (c.1500 onwards).
New sub-clades of haplogroups may be discovered when an
individual tests, especially if they are non-European. Most significant
of these new discoveries was in 2013 when the haplogroup A00
was discovered, which required theories about Y-chromosomal Adam to be
significantly revised. The haplogroup was discovered when an
African-American man tested STRs at FamilyTreeDNA and his results were
found to be unusual. SNP testing confirmed that he does not descend
patrilineally from the "old" Y-chromosomal Adam and so a much older man
became Y-Chromosomal Adam.
Using DNA test results
Ethnicity estimates
Many
companies offer a percentage breakdown by ethnicity or region.
Generally the world is specified into about 20–25 regions, and the
approximate percentage of DNA inherited from each is stated. This is
usually done by comparing the frequency of each Autosomal DNA marker tested to many population groups.
The reliability of this type of test is dependent on comparative
population size, the number of markers tested, the ancestry informative
value of the SNPs tested, and the degree of admixture in the person
tested. Earlier ethnicity estimates were often wildly inaccurate, but as
companies receive more samples over time, ethnicity estimates have
become more accurate. Testing companies such as Ancestry.com
will often regularly update their ethnicity estimates, which has caused
some controversy from customers as their results update.
Usually the results at the continental level are accurate, but more
specific assertions of the test may turn out to be incorrect.
Audience
The
interest in genealogical DNA tests has been linked to both an increase
in curiosity about traditional genealogy and to more general personal
origins. Those who test for traditional genealogy often utilize a
combination of autosomal, mitochondrial, and Y-Chromosome tests. Those
with an interest in personal ethnic origins are more likely to use an
autosomal test. However, answering specific questions about the ethnic
origins of a particular lineage may be best suited to an mtDNA test or a
Y-DNA test.
Maternal origin tests
For
recent genealogy, exact matching on the mtDNA full sequence is used to
confirm a common ancestor on the direct maternal line between two
suspected relatives. Because mtDNA mutations are very rare, a nearly perfect match is not usually considered relevant to the most recent 1 to 16 generations. In cultures lacking matrilineal surnames
to pass down, neither relative above is likely to have as many
generations of ancestors in their matrilineal information table as in
the above patrilineal or Y-DNA case: for further information on this
difficulty in traditional genealogy, due to lack of matrilineal surnames (or matrinames), see Matriname.
However, the foundation of testing is still two suspected descendants
of one person. This hypothesize and test DNA pattern is the same one
used for autosomal DNA and Y-DNA.
Tests for ethnicity and membership of other groups
European genetic structure (based on Autosomal SNPs) by PCA
As discussed above, autosomal tests usually report the ethnic
proportions of the individual. These attempt to measure an individual's
mixed geographic heritage by identifying particular markers, called
ancestry informative markers or AIM, that are associated with
populations of specific geographical areas. Geneticist Adam Rutherford
has written that these tests "don’t necessarily show your geographical
origins in the past. They show with whom you have common ancestry
today."
The haplogroups determined by Y-DNA and mtDNA tests are often
unevenly geographically distributed. Many direct-to-consumer DNA tests
described this association to infer the test-taker's ancestral homeland. Most tests describe haplogroups according to their most frequently associated continent (e.g., a "European haplogroup").
When Leslie Emery and collaborators performed a trial of mtDNA
haplogroups as a predictor of continental origin on individuals in the
Human Genetic Diversity Panel (HGDP) and 1000 Genomes (1KGP) datasets,
they found that only 14 of 23 haplogroups had a success rate above 50%
among the HGDP samples, as did "about half" of the haplogroups in the
1KGP.
The authors concluded that, for most people, "mtDNA-haplogroup
membership provides limited information about either continental
ancestry or continental region of origin."
African ancestry
Y-DNA and mtDNA testing may be able to determine with which peoples in present-day Africa
a person shares a direct line of part of his or her ancestry, but
patterns of historic migration and historical events cloud the tracing
of ancestral groups. Due to joint long histories in the US,
approximately 30% of African American males have a European Y-Chromosome haplogroup
Approximately 58% of African Americans have at least the equivalent of
one great-grandparent (13%) of European ancestry. Only about 5% have the
equivalent of one great-grandparent of Native American ancestry. By
the early 19th century, substantial families of Free Persons of Color
had been established in the Chesapeake Bay area who were descended from
free people during the colonial period; most of those have been
documented as descended from white men and African women (servant, slave
or free). Over time various groups married more within mixed-race,
black or white communities.
According to authorities like Salas, nearly three-quarters of the ancestors of African Americans taken in slavery
came from regions of West Africa. The African-American movement to
discover and identify with ancestral tribes has burgeoned since DNA
testing became available. African Americans usually cannot easily trace
their ancestry during the years of slavery through surname research, census and property records, and other traditional means. Genealogical DNA testing may provide a tie to regional African heritage.
United States – Melungeon testing
Melungeons
are one of numerous multiracial groups in the United States with
origins wrapped in myth. The historical research of Paul Heinegg has
documented that many of the Melungeon groups in the Upper South were
descended from mixed-race people who were free in colonial Virginia and
the result of unions between the Europeans and Africans. They moved to
the frontiers of Virginia, North Carolina, Kentucky and Tennessee to
gain some freedom from the racial barriers of the plantation areas.
Several efforts, including a number of ongoing studies, have examined
the genetic makeup of families historically identified as Melungeon.
Most results point primarily to a mixture of European and African, which
is supported by historical documentation. Some may have Native American
heritage as well. Though some companies provide additional Melungeon
research materials with Y-DNA and mtDNA tests, any test will allow
comparisons with the results of current and past Melungeon DNA studies
Native American ancestry
The pre-columbian indigenous people of the United States are called "Native Americans" in American English. Autosomal testing, Y-DNA, and mtDNA testing can be conducted to determine the ancestry of Native Americans. A mitochondrial Haplogroup determination test based on mutations in Hypervariable Region 1 and 2 may establish whether a person's direct female line belongs to one of the canonical Native American Haplogroups, A, B, C, D or X. The vast majority of Native American individuals belong to one of the five identified mtDNA Haplogroups.
Thus, being in one of those groups provides evidence of potential
Native American descent. However, DNA ethnicity results cannot be used
as a substitute for legal documentation. Native American tribes
have their own requirements for membership, often based on at least one
of a person's ancestors having been included on tribal-specific Native
American censuses (or final rolls) prepared during treaty-making, relocation to reservations or apportionment of land in the late 19th century and early 20th century. One example is the Dawes Rolls.
Cohanim ancestry
The Cohanim (or Kohanim) is a patrilineal priestly line of descent in Judaism. According to the Bible, the ancestor of the Cohanim is Aaron, brother of Moses.
Many believe that descent from Aaron is verifiable with a Y-DNA test:
the first published study in genealogical Y-Chromosome DNA testing found
that a significant percentage of Cohens had distinctively similar DNA,
rather more so than general Jewish or Middle Eastern populations. These
Cohens tended to belong to Haplogroup J, with Y-STR values clustered unusually closely around a haplotype known as the Cohen Modal Haplotype
(CMH). This could be consistent with a shared common ancestor, or with
the hereditary priesthood having originally been founded from members of
a single closely related clan.
Nevertheless, the original studies tested only six Y-STR markers,
which is considered a low-resolution test. In response to the low
resolution of the original 6-marker CMH, the testing company FTDNA
released a 12-marker CMH signature that was more specific to the large
closely related group of Cohens in Haplogroup J1.
A further academic study published in 2009 examined more STR markers and identified a more sharply defined SNP haplogroup, J1e*
(now J1c3, also called J-P58*) for the J1 lineage. The research found
"that 46.1% of Kohanim carry Y chromosomes belonging to a single
paternal lineage (J-P58*) that likely originated in the Near East well
before the dispersal of Jewish groups in the Diaspora. Support for a
Near Eastern origin of this lineage comes from its high frequency in our
sample of Bedouins, Yemenis (67%), and Jordanians (55%) and its
precipitous drop in frequency as one moves away from Saudi Arabia and
the Near East (Fig. 4). Moreover, there is a striking contrast between
the relatively high frequency of J-58* in Jewish populations (»20%) and
Kohanim (»46%) and its vanishingly low frequency in our sample of
non-Jewish populations that hosted Jewish diaspora communities outside
of the Near East."
Recent phylogenetic research for haplogroup J-M267 placed the
"Y-chromosomal Aaron" in a subhaplogroup of J-L862, L147.1 (age estimate
5631-6778yBP yBP):
YSC235>PF4847/CTS11741>YSC234>ZS241>ZS227>Z18271 (age
estimate 2731yBP).
Benefits
Genealogical DNA tests have become popular due to the ease of testing at home and their usefulness in supplementing genealogical research.
Genealogical DNA tests allow for an individual to determine with high
accuracy whether he or she is related to another person within a certain
time frame, or with certainty that he or she is not related. DNA tests
are perceived as more scientific, conclusive and expeditious than
searching the civil records. However, they are limited by restrictions
on lines that may be studied. The civil records are always only as
accurate as the individuals having provided or written the information.
Y-DNA testing
results are normally stated as probabilities: For example, with the
same surname a perfect 37/37 marker test match gives a 95% likelihood of
the most recent common ancestor (MRCA) being within 8 generations, while a 111 of 111 marker match gives the same 95% likelihood of the MRCA being within only 5 generations back.
As presented above in mtDNA testing,
if a perfect match is found, the mtDNA test results can be helpful. In
some cases, research according to traditional genealogy methods
encounters difficulties due to the lack of regularly recorded
matrilineal surname information in many cultures (see Matrilineal surname).
Autosomal DNA combined with genealogical research has been used by adoptees to find their biological parents, has been used to find the name and family of unidentified bodies and by law enforcement agencies to apprehend criminals (for example, the Contra Costa County District Attorney's office used the "open-source" genetic genealogy site GEDmatch to find relatives of the suspect in the Golden State Killer case.). The Atlantic
magazine commented in 2018 that "Now, the floodgates are open. ..a
small, volunteer-run website, GEDmatch.com, has become ... the de facto
DNA and genealogy database for all of law enforcement." Family Tree DNA announced in February 2019 it was allowing the FBI to access its DNA data for cases of murder and rape. However in May 2019 GEDmatch initiated stricter rules for accessing their autosomal DNA database
and Family Tree DNA shut down their Y-DNA database ysearch.org, making
it more difficult for law enforcement agencies to solve cases.
Drawbacks
Common concerns about genealogical DNA testing are cost and privacy issues. Some testing companies retain samples and results for their own use without a privacy agreement with subjects.
Autosomal DNA tests can identify relationships but they can be misinterpreted.
For example, transplants of stem cell or bone marrow will produce
matches with the donor. In addition, identical twins (who have identical
DNA) can give unexpected results.
Testing of the Y-DNA lineage from father to son may reveal complications, due to unusual mutations, secret adoptions, and non-paternity events (i.e., that the perceived father in a generation is not the father indicated by written birth records). According to the Ancestry and Ancestry Testing Task Force of the American Society of Human Genetics, autosomal tests cannot detect "large portions" of DNA from distant ancestors because it has not been inherited.
With the increasing popularity of the use of DNA tests for
ethnicity tests, uncertainties and errors in ethnicity estimates are a
drawback for Genetic genealogy. While ethnicity estimates at the
continental level should be accurate (with the possible exception of
East Asia and the Americas), sub-continental estimates, especially in
Europe, are often inaccurate. Customers may be misinformed about the
uncertainties and errors of the estimates.
Some have recommended government or other regulation of ancestry testing to ensure its performance to an agreed standard.
A number of law enforcement agencies took legal action to compel
genetic genealogy companies to release genetic information that could match cold case crime victims or perpetrators. A number of companies fought the requests.
Medical information
Though
genealogical DNA tests are not designed mainly for medical purposes,
autosomal DNA tests can be used to analyze the probability of hundreds
of heritable medical conditions,
albeit the result is complex to understand and may confuse a
non-expert. 23andMe provides medical and trait information from their
genealogical DNA test and for a fee the Promethease web site analyses genealogical DNA test data from Family Tree DNA, 23andMe, or AncestryDNA for medical information.
Promethease, and its research paper crawling database SNPedia, has
received criticism for technical complexity and a poorly defined
"magnitude" scale that causes misconceptions, confusion and panic among
its users.
The testing of full MtDNA and YDNA sequences is still somewhat
controversial as it may reveal even more medical information. For
example, a correlation exists between a lack of Y-DNA marker DYS464 and infertility, and between mtDNA haplogroup H and protection from sepsis. Certain haplogroups have been linked to longevity in some population groups.
The field of linkage disequilibrium, unequal association of genetic
disorders with a certain mitochondrial lineage, is in its infancy, but
those mitochondrial mutations that have been linked are searchable in
the genome database Mitomap. Family Tree DNA's MtFull Sequence test analyses the full MtDNA genome and the National Human Genome Research Institute operates the Genetic And Rare Disease Information Center that can assist consumers in identifying an appropriate screening test and help locate a nearby medical center that offers such a test.
DNA in genealogy software
Some genealogy software
programs — such as Family Tree Maker, Legacy Family Tree (Deluxe
Edition) and the Swedish program Genney — allow recording DNA marker
test results. This allows for tracking of both Y-chromosome and mtDNA
tests, and recording results for relatives.