| Genetic engineering |
|---|
 |
| Genetically modified organisms |
| History and regulation |
| Process |
| Applications |
| Controversies |
Genetic engineering, also called genetic modification or genetic manipulation, is the direct manipulation of an organism's genes using biotechnology. It is a set of technologies used to change the genetic makeup of cells, including the transfer of genes within and across species boundaries to produce improved or novel organisms. New DNA is obtained by either isolating and copying the genetic material of interest using recombinant DNA methods or by artificially synthesising the DNA. A construct is usually created and used to insert this DNA into the host organism. The first recombinant DNA molecule was made by Paul Berg in 1972 by combining DNA from the monkey virus SV40 with the lambda virus. As well as inserting genes, the process can be used to remove, or "knock out", genes. The new DNA can be inserted randomly, or targeted to a specific part of the genome.
An organism that is generated through genetic engineering is considered to be genetically modified (GM) and the resulting entity is a genetically modified organism (GMO). The first GMO was a bacterium generated by Herbert Boyer and Stanley Cohen in 1973. Rudolf Jaenisch created the first GM animal when he inserted foreign DNA into a mouse in 1974. The first company to focus on genetic engineering, Genentech, was founded in 1976 and started the production of human proteins. Genetically engineered human insulin was produced in 1978 and insulin-producing bacteria were commercialised in 1982. Genetically modified food has been sold since 1994, with the release of the Flavr Savr tomato. The Flavr Savr was engineered to have a longer shelf life, but most current GM crops are modified to increase resistance to insects and herbicides. GloFish, the first GMO designed as a pet, was sold in the United States in December 2003. In 2016 salmon modified with a growth hormone were sold.
Genetic engineering has been applied in numerous fields including research, medicine, industrial biotechnology and agriculture. In research GMOs are used to study gene function and expression through loss of function, gain of function, tracking and expression experiments. By knocking out genes responsible for certain conditions it is possible to create animal model organisms of human diseases. As well as producing hormones, vaccines and other drugs, genetic engineering has the potential to cure genetic diseases through gene therapy. The same techniques that are used to produce drugs can also have industrial applications such as producing enzymes for laundry detergent, cheeses and other products.
The rise of commercialised genetically modified crops has provided economic benefit to farmers in many different countries, but has also been the source of most of the controversy surrounding the technology. This has been present since its early use; the first field trials were destroyed by anti-GM activists. Although there is a scientific consensus that currently available food derived from GM crops poses no greater risk to human health than conventional food, GM food safety is a leading concern with critics. Gene flow, impact on non-target organisms, control of the food supply and intellectual property rights have also been raised as potential issues. These concerns have led to the development of a regulatory framework, which started in 1975. It has led to an international treaty, the Cartagena Protocol on Biosafety, that was adopted in 2000. Individual countries have developed their own regulatory systems regarding GMOs, with the most marked differences occurring between the US and Europe.
Overview
Genetic engineering is a process that alters the genetic structure of an organism by either removing or introducing DNA. Unlike traditional animal and plant breeding, which involves doing multiple crosses and then selecting for the organism with the desired phenotype, genetic engineering takes the gene directly from one organism and delivers it to the other. This is much faster, can be used to insert any genes from any organism (even ones from different domains) and prevents other undesirable genes from also being added.
Genetic engineering could potentially fix severe genetic disorders in humans by replacing the defective gene with a functioning one. It is an important tool in research that allows the function of specific genes to be studied. Drugs, vaccines and other products have been harvested from organisms engineered to produce them. Crops have been developed that aid food security by increasing yield, nutritional value and tolerance to environmental stresses.
The DNA can be introduced directly into the host organism or into a cell that is then fused or hybridised with the host. This relies on recombinant nucleic acid techniques to form new combinations of heritable genetic material followed by the incorporation of that material either indirectly through a vector system or directly through micro-injection, macro-injection or micro-encapsulation.
Genetic engineering does not normally include traditional breeding, in vitro fertilisation, induction of polyploidy, mutagenesis and cell fusion techniques that do not use recombinant nucleic acids or a genetically modified organism in the process. However, some broad definitions of genetic engineering include selective breeding. Cloning and stem cell research, although not considered genetic engineering, are closely related and genetic engineering can be used within them. Synthetic biology is an emerging discipline that takes genetic engineering a step further by introducing artificially synthesised material into an organism. Such synthetic DNA as Artificially Expanded Genetic Information System and Hachimoji DNA is made in this new field.
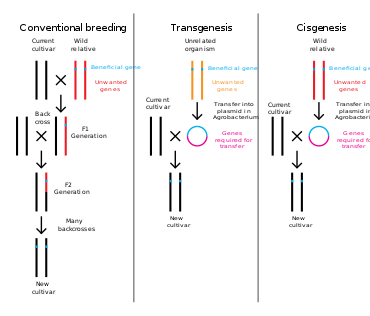
Plants, animals or microorganisms that have been changed through genetic engineering are termed genetically modified organisms or GMOs. If genetic material from another species is added to the host, the resulting organism is called transgenic. If genetic material from the same species or a species that can naturally breed with the host is used the resulting organism is called cisgenic. If genetic engineering is used to remove genetic material from the target organism the resulting organism is termed a knockout organism. In Europe genetic modification is synonymous with genetic engineering while within the United States of America and Canada genetic modification can also be used to refer to more conventional breeding methods.
History
Humans have altered the genomes of species for thousands of years through selective breeding, or artificial selection as contrasted with natural selection. More recently, mutation breeding has used exposure to chemicals or radiation to produce a high frequency of random mutations, for selective breeding purposes. Genetic engineering as the direct manipulation of DNA by humans outside breeding and mutations has only existed since the 1970s. The term "genetic engineering" was first coined by Jack Williamson in his science fiction novel Dragon's Island, published in 1951 – one year before DNA's role in heredity was confirmed by Alfred Hershey and Martha Chase, and two years before James Watson and Francis Crick showed that the DNA molecule has a double-helix structure – though the general concept of direct genetic manipulation was explored in rudimentary form in Stanley G. Weinbaum's 1936 science fiction story Proteus Island.
In 1972, Paul Berg created the first recombinant DNA molecules by combining DNA from the monkey virus SV40 with that of the lambda virus. In 1973 Herbert Boyer and Stanley Cohen created the first transgenic organism by inserting antibiotic resistance genes into the plasmid of an Escherichia coli bacterium. A year later Rudolf Jaenisch created a transgenic mouse by introducing foreign DNA into its embryo, making it the world's first transgenic animal These achievements led to concerns in the scientific community about potential risks from genetic engineering, which were first discussed in depth at the Asilomar Conference in 1975. One of the main recommendations from this meeting was that government oversight of recombinant DNA research should be established until the technology was deemed safe.
In 1976 Genentech, the first genetic engineering company, was founded by Herbert Boyer and Robert Swanson and a year later the company produced a human protein (somatostatin) in E.coli. Genentech announced the production of genetically engineered human insulin in 1978. In 1980, the U.S. Supreme Court in the Diamond v. Chakrabarty case ruled that genetically altered life could be patented. The insulin produced by bacteria was approved for release by the Food and Drug Administration (FDA) in 1982.
In 1983, a biotech company, Advanced Genetic Sciences (AGS) applied for U.S. government authorisation to perform field tests with the ice-minus strain of Pseudomonas syringae to protect crops from frost, but environmental groups and protestors delayed the field tests for four years with legal challenges. In 1987, the ice-minus strain of P. syringae became the first genetically modified organism (GMO) to be released into the environment when a strawberry field and a potato field in California were sprayed with it. Both test fields were attacked by activist groups the night before the tests occurred: "The world's first trial site attracted the world's first field trasher".
The first field trials of genetically engineered plants occurred in France and the US in 1986, tobacco plants were engineered to be resistant to herbicides. The People's Republic of China was the first country to commercialise transgenic plants, introducing a virus-resistant tobacco in 1992. In 1994 Calgene attained approval to commercially release the first genetically modified food, the Flavr Savr, a tomato engineered to have a longer shelf life. In 1994, the European Union approved tobacco engineered to be resistant to the herbicide bromoxynil, making it the first genetically engineered crop commercialised in Europe. In 1995, Bt Potato was approved safe by the Environmental Protection Agency, after having been approved by the FDA, making it the first pesticide producing crop to be approved in the US. In 2009 11 transgenic crops were grown commercially in 25 countries, the largest of which by area grown were the US, Brazil, Argentina, India, Canada, China, Paraguay and South Africa.
In 2010, scientists at the J. Craig Venter Institute created the first synthetic genome and inserted it into an empty bacterial cell. The resulting bacterium, named Mycoplasma laboratorium, could replicate and produce proteins. Four years later this was taken a step further when a bacterium was developed that replicated a plasmid containing a unique base pair, creating the first organism engineered to use an expanded genetic alphabet. In 2012, Jennifer Doudna and Emmanuelle Charpentier collaborated to develop the CRISPR/Cas9 system, a technique which can be used to easily and specifically alter the genome of almost any organism.
Process
Creating a GMO is a multi-step process. Genetic engineers must first choose what gene they wish to insert into the organism. This is driven by what the aim is for the resultant organism and is built on earlier research. Genetic screens can be carried out to determine potential genes and further tests then used to identify the best candidates. The development of microarrays, transcriptomics and genome sequencing has made it much easier to find suitable genes. Luck also plays its part; the round-up ready gene was discovered after scientists noticed a bacterium thriving in the presence of the herbicide.
Gene isolation and cloning
The next step is to isolate the candidate gene. The cell containing the gene is opened and the DNA is purified. The gene is separated by using restriction enzymes to cut the DNA into fragments or polymerase chain reaction (PCR) to amplify up the gene segment. These segments can then be extracted through gel electrophoresis. If the chosen gene or the donor organism's genome has been well studied it may already be accessible from a genetic library. If the DNA sequence is known, but no copies of the gene are available, it can also be artificially synthesised. Once isolated the gene is ligated into a plasmid that is then inserted into a bacterium. The plasmid is replicated when the bacteria divide, ensuring unlimited copies of the gene are available.
Before the gene is inserted into the target organism it must be combined with other genetic elements. These include a promoter and terminator region, which initiate and end transcription. A selectable marker gene is added, which in most cases confers antibiotic resistance, so researchers can easily determine which cells have been successfully transformed. The gene can also be modified at this stage for better expression or effectiveness. These manipulations are carried out using recombinant DNA techniques, such as restriction digests, ligations and molecular cloning.
Inserting DNA into the host genome
There are a number of techniques used to insert genetic material into the host genome. Some bacteria can naturally take up foreign DNA. This ability can be induced in other bacteria via stress (e.g. thermal or electric shock), which increases the cell membrane's permeability to DNA; up-taken DNA can either integrate with the genome or exist as extrachromosomal DNA. DNA is generally inserted into animal cells using microinjection, where it can be injected through the cell's nuclear envelope directly into the nucleus, or through the use of viral vectors.
Plant genomes can be engineered by physical methods or by use of Agrobacterium for the delivery of sequences hosted in T-DNA binary vectors. In plants the DNA is often inserted using Agrobacterium-mediated transformation, taking advantage of the Agrobacteriums T-DNA sequence that allows natural insertion of genetic material into plant cells. Other methods include biolistics, where particles of gold or tungsten are coated with DNA and then shot into young plant cells, and electroporation, which involves using an electric shock to make the cell membrane permeable to plasmid DNA.
As only a single cell is transformed with genetic material, the organism must be regenerated from that single cell. In plants this is accomplished through the use of tissue culture. In animals it is necessary to ensure that the inserted DNA is present in the embryonic stem cells. Bacteria consist of a single cell and reproduce clonally so regeneration is not necessary. Selectable markers are used to easily differentiate transformed from untransformed cells. These markers are usually present in the transgenic organism, although a number of strategies have been developed that can remove the selectable marker from the mature transgenic plant.
Further testing using PCR, Southern hybridization, and DNA sequencing is conducted to confirm that an organism contains the new gene. These tests can also confirm the chromosomal location and copy number of the inserted gene. The presence of the gene does not guarantee it will be expressed at appropriate levels in the target tissue so methods that look for and measure the gene products (RNA and protein) are also used. These include northern hybridisation, quantitative RT-PCR, Western blot, immunofluorescence, ELISA and phenotypic analysis.
The new genetic material can be inserted randomly within the host genome or targeted to a specific location. The technique of gene targeting uses homologous recombination to make desired changes to a specific endogenous gene. This tends to occur at a relatively low frequency in plants and animals and generally requires the use of selectable markers. The frequency of gene targeting can be greatly enhanced through genome editing. Genome editing uses artificially engineered nucleases that create specific double-stranded breaks at desired locations in the genome, and use the cell's endogenous mechanisms to repair the induced break by the natural processes of homologous recombination and nonhomologous end-joining. There are four families of engineered nucleases: meganucleases, zinc finger nucleases, transcription activator-like effector nucleases (TALENs), and the Cas9-guideRNA system (adapted from CRISPR). TALEN and CRISPR are the two most commonly used and each has its own advantages. TALENs have greater target specificity, while CRISPR is easier to design and more efficient. In addition to enhancing gene targeting, engineered nucleases can be used to introduce mutations at endogenous genes that generate a gene knockout.
Applications
Genetic engineering has applications in medicine, research, industry and agriculture and can be used on a wide range of plants, animals and microorganisms. Bacteria, the first organisms to be genetically modified, can have plasmid DNA inserted containing new genes that code for medicines or enzymes that process food and other substrates. Plants have been modified for insect protection, herbicide resistance, virus resistance, enhanced nutrition, tolerance to environmental pressures and the production of edible vaccines. Most commercialised GMOs are insect resistant or herbicide tolerant crop plants. Genetically modified animals have been used for research, model animals and the production of agricultural or pharmaceutical products. The genetically modified animals include animals with genes knocked out, increased susceptibility to disease, hormones for extra growth and the ability to express proteins in their milk.
Medicine
Genetic engineering has many applications to medicine that include the manufacturing of drugs, creation of model animals that mimic human conditions and gene therapy. One of the earliest uses of genetic engineering was to mass-produce human insulin in bacteria. This application has now been applied to human growth hormones, follicle stimulating hormones (for treating infertility), human albumin, monoclonal antibodies, antihemophilic factors, vaccines and many other drugs. Mouse hybridomas, cells fused together to create monoclonal antibodies, have been adapted through genetic engineering to create human monoclonal antibodies. In 2017, genetic engineering of chimeric antigen receptors on a patient's own T-cells was approved by the U.S. FDA as a treatment for the cancer acute lymphoblastic leukemia. Genetically engineered viruses are being developed that can still confer immunity, but lack the infectious sequences.
Genetic engineering is also used to create animal models of human diseases. Genetically modified mice are the most common genetically engineered animal model. They have been used to study and model cancer (the oncomouse), obesity, heart disease, diabetes, arthritis, substance abuse, anxiety, aging and Parkinson disease. Potential cures can be tested against these mouse models. Also genetically modified pigs have been bred with the aim of increasing the success of pig to human organ transplantation.
Gene therapy is the genetic engineering of humans, generally by replacing defective genes with effective ones. Clinical research using somatic gene therapy has been conducted with several diseases, including X-linked SCID, chronic lymphocytic leukemia (CLL), and Parkinson's disease. In 2012, Alipogene tiparvovec became the first gene therapy treatment to be approved for clinical use. In 2015 a virus was used to insert a healthy gene into the skin cells of a boy suffering from a rare skin disease, epidermolysis bullosa, in order to grow, and then graft healthy skin onto 80 percent of the boy's body which was affected by the illness.
Germline gene therapy would result in any change being inheritable, which has raised concerns within the scientific community. In 2015, CRISPR was used to edit the DNA of non-viable human embryos, leading scientists of major world academies to call for a moratorium on inheritable human genome edits. There are also concerns that the technology could be used not just for treatment, but for enhancement, modification or alteration of a human beings' appearance, adaptability, intelligence, character or behavior. The distinction between cure and enhancement can also be difficult to establish. In November 2018, He Jiankui announced that he had edited the genomes of two human embryos, to attempt to disable the CCR5 gene, which codes for a receptor that HIV uses to enter cells. He said that twin girls, Lulu and Nana, had been born a few weeks earlier. He said that the girls still carried functional copies of CCR5 along with disabled CCR5 (mosaicism) and were still vulnerable to HIV. The work was widely condemned as unethical, dangerous, and premature. Currently, germline modification is banned in 40 countries. Scientists that do this type of research will often let embryos grow for a few days without allowing it to develop into a baby.
Researchers are altering the genome of pigs to induce the growth of human organs to be used in transplants. Scientists are creating "gene drives", changing the genomes of mosquitoes to make them immune to malaria, and then looking to spread the genetically altered mosquitoes throughout the mosquito population in the hopes of eliminating the disease.
Research
Genetic engineering is an important tool for natural scientists, with the creation of transgenic organisms one of the most important tools for analysis of gene function. Genes and other genetic information from a wide range of organisms can be inserted into bacteria for storage and modification, creating genetically modified bacteria in the process. Bacteria are cheap, easy to grow, clonal, multiply quickly, relatively easy to transform and can be stored at -80 °C almost indefinitely. Once a gene is isolated it can be stored inside the bacteria providing an unlimited supply for research. Organisms are genetically engineered to discover the functions of certain genes. This could be the effect on the phenotype of the organism, where the gene is expressed or what other genes it interacts with. These experiments generally involve loss of function, gain of function, tracking and expression.
- Loss of function experiments, such as in a gene knockout experiment, in which an organism is engineered to lack the activity of one or more genes. In a simple knockout a copy of the desired gene has been altered to make it non-functional. Embryonic stem cells incorporate the altered gene, which replaces the already present functional copy. These stem cells are injected into blastocysts, which are implanted into surrogate mothers. This allows the experimenter to analyse the defects caused by this mutation and thereby determine the role of particular genes. It is used especially frequently in developmental biology. When this is done by creating a library of genes with point mutations at every position in the area of interest, or even every position in the whole gene, this is called "scanning mutagenesis". The simplest method, and the first to be used, is "alanine scanning", where every position in turn is mutated to the unreactive amino acid alanine.
- Gain of function experiments, the logical counterpart of knockouts. These are sometimes performed in conjunction with knockout experiments to more finely establish the function of the desired gene. The process is much the same as that in knockout engineering, except that the construct is designed to increase the function of the gene, usually by providing extra copies of the gene or inducing synthesis of the protein more frequently. Gain of function is used to tell whether or not a protein is sufficient for a function, but does not always mean it's required, especially when dealing with genetic or functional redundancy.
- Tracking experiments, which seek to gain information about the localisation and interaction of the desired protein. One way to do this is to replace the wild-type gene with a 'fusion' gene, which is a juxtaposition of the wild-type gene with a reporting element such as green fluorescent protein (GFP) that will allow easy visualisation of the products of the genetic modification. While this is a useful technique, the manipulation can destroy the function of the gene, creating secondary effects and possibly calling into question the results of the experiment. More sophisticated techniques are now in development that can track protein products without mitigating their function, such as the addition of small sequences that will serve as binding motifs to monoclonal antibodies.
- Expression studies aim to discover where and when specific proteins are produced. In these experiments, the DNA sequence before the DNA that codes for a protein, known as a gene's promoter, is reintroduced into an organism with the protein coding region replaced by a reporter gene such as GFP or an enzyme that catalyses the production of a dye. Thus the time and place where a particular protein is produced can be observed. Expression studies can be taken a step further by altering the promoter to find which pieces are crucial for the proper expression of the gene and are actually bound by transcription factor proteins; this process is known as promoter bashing.
Industrial
Organisms can have their cells transformed with a gene coding for a useful protein, such as an enzyme, so that they will overexpress the desired protein. Mass quantities of the protein can then be manufactured by growing the transformed organism in bioreactor equipment using industrial fermentation, and then purifying the protein. Some genes do not work well in bacteria, so yeast, insect cells or mammalians cells can also be used. These techniques are used to produce medicines such as insulin, human growth hormone, and vaccines, supplements such as tryptophan, aid in the production of food (chymosin in cheese making) and fuels. Other applications with genetically engineered bacteria could involve making them perform tasks outside their natural cycle, such as making biofuels, cleaning up oil spills, carbon and other toxic waste and detecting arsenic in drinking water. Certain genetically modified microbes can also be used in biomining and bioremediation, due to their ability to extract heavy metals from their environment and incorporate them into compounds that are more easily recoverable.
In materials science, a genetically modified virus has been used in a research laboratory as a scaffold for assembling a more environmentally friendly lithium-ion battery. Bacteria have also been engineered to function as sensors by expressing a fluorescent protein under certain environmental conditions.
Agriculture
One of the best-known and controversial applications of genetic engineering is the creation and use of genetically modified crops or genetically modified livestock to produce genetically modified food. Crops have been developed to increase production, increase tolerance to abiotic stresses, alter the composition of the food, or to produce novel products.
The first crops to be released commercially on a large scale provided protection from insect pests or tolerance to herbicides. Fungal and virus resistant crops have also been developed or are in development. This makes the insect and weed management of crops easier and can indirectly increase crop yield. GM crops that directly improve yield by accelerating growth or making the plant more hardy (by improving salt, cold or drought tolerance) are also under development. In 2016 Salmon have been genetically modified with growth hormones to reach normal adult size much faster.
GMOs have been developed that modify the quality of produce by increasing the nutritional value or providing more industrially useful qualities or quantities. The Amflora potato produces a more industrially useful blend of starches. Soybeans and canola have been genetically modified to produce more healthy oils. The first commercialised GM food was a tomato that had delayed ripening, increasing its shelf life.
Plants and animals have been engineered to produce materials they do not normally make. Pharming uses crops and animals as bioreactors to produce vaccines, drug intermediates, or the drugs themselves; the useful product is purified from the harvest and then used in the standard pharmaceutical production process. Cows and goats have been engineered to express drugs and other proteins in their milk, and in 2009 the FDA approved a drug produced in goat milk.
Other applications
Genetic engineering has potential applications in conservation and natural area management. Gene transfer through viral vectors has been proposed as a means of controlling invasive species as well as vaccinating threatened fauna from disease. Transgenic trees have been suggested as a way to confer resistance to pathogens in wild populations. With the increasing risks of maladaptation in organisms as a result of climate change and other perturbations, facilitated adaptation through gene tweaking could be one solution to reducing extinction risks. Applications of genetic engineering in conservation are thus far mostly theoretical and have yet to be put into practice.
Genetic engineering is also being used to create microbial art. Some bacteria have been genetically engineered to create black and white photographs. Novelty items such as lavender-colored carnations, blue roses, and glowing fish have also been produced through genetic engineering.
Regulation
The regulation of genetic engineering concerns the approaches taken by governments to assess and manage the risks associated with the development and release of GMOs. The development of a regulatory framework began in 1975, at Asilomar, California. The Asilomar meeting recommended a set of voluntary guidelines regarding the use of recombinant technology. As the technology improved the US established a committee at the Office of Science and Technology, which assigned regulatory approval of GM food to the USDA, FDA and EPA. The Cartagena Protocol on Biosafety, an international treaty that governs the transfer, handling, and use of GMOs, was adopted on 29 January 2000. One hundred and fifty-seven countries are members of the Protocol and many use it as a reference point for their own regulations.
The legal and regulatory status of GM foods varies by country, with some nations banning or restricting them, and others permitting them with widely differing degrees of regulation. Some countries allow the import of GM food with authorisation, but either do not allow its cultivation (Russia, Norway, Israel) or have provisions for cultivation even though no GM products are yet produced (Japan, South Korea). Most countries that do not allow GMO cultivation do permit research. Some of the most marked differences occurring between the US and Europe. The US policy focuses on the product (not the process), only looks at verifiable scientific risks and uses the concept of substantial equivalence. The European Union by contrast has possibly the most stringent GMO regulations in the world. All GMOs, along with irradiated food, are considered "new food" and subject to extensive, case-by-case, science-based food evaluation by the European Food Safety Authority. The criteria for authorisation fall in four broad categories: "safety", "freedom of choice", "labelling", and "traceability". The level of regulation in other countries that cultivate GMOs lie in between Europe and the United States.
| Region | Regulators | Notes |
|---|---|---|
| US | USDA, FDA and EPA |
|
| Europe | European Food Safety Authority |
|
| Canada | Health Canada and the Canadian Food Inspection Agency | Regulated products with novel features regardless of method of origin |
| Africa | Common Market for Eastern and Southern Africa | Final decision lies with each individual country. |
| China | Office of Agricultural Genetic Engineering Biosafety Administration |
|
| India | Institutional Biosafety Committee, Review Committee on Genetic Manipulation and Genetic Engineering Approval Committee |
|
| Argentina | National Agricultural Biotechnology Advisory Committee (environmental impact), the National Service of Health and Agrifood Quality (food safety) and the National Agribusiness Direction (effect on trade) | Final decision made by the Secretariat of Agriculture, Livestock, Fishery and Food. |
| Brazil | National Biosafety Technical Commission (environmental and food safety) and the Council of Ministers (commercial and economical issues) |
|
| Australia | Office of the Gene Technology Regulator (oversees all GM products), Therapeutic Goods Administration (GM medicines) and Food Standards Australia New Zealand (GM food). | The individual state governments can then assess the impact of release on markets and trade and apply further legislation to control approved genetically modified products. |
One of the key issues concerning regulators is whether GM products should be labeled. The European Commission says that mandatory labeling and traceability are needed to allow for informed choice, avoid potential false advertising and facilitate the withdrawal of products if adverse effects on health or the environment are discovered. The American Medical Association and the American Association for the Advancement of Science say that absent scientific evidence of harm even voluntary labeling is misleading and will falsely alarm consumers. Labeling of GMO products in the marketplace is required in 64 countries. Labeling can be mandatory up to a threshold GM content level (which varies between countries) or voluntary. In Canada and the US labeling of GM food is voluntary, while in Europe all food (including processed food) or feed which contains greater than 0.9% of approved GMOs must be labelled.
Controversy
Critics have objected to the use of genetic engineering on several grounds, including ethical, ecological and economic concerns. Many of these concerns involve GM crops and whether food produced from them is safe and what impact growing them will have on the environment. These controversies have led to litigation, international trade disputes, and protests, and to restrictive regulation of commercial products in some countries.
Accusations that scientists are "playing God" and other religious issues have been ascribed to the technology from the beginning. Other ethical issues raised include the patenting of life, the use of intellectual property rights, the level of labeling on products, control of the food supply and the objectivity of the regulatory process. Although doubts have been raised, economically most studies have found growing GM crops to be beneficial to farmers.
Gene flow between GM crops and compatible plants, along with increased use of selective herbicides, can increase the risk of "superweeds" developing. Other environmental concerns involve potential impacts on non-target organisms, including soil microbes, and an increase in secondary and resistant insect pests. Many of the environmental impacts regarding GM crops may take many years to be understood and are also evident in conventional agriculture practices. With the commercialisation of genetically modified fish there are concerns over what the environmental consequences will be if they escape.
There are three main concerns over the safety of genetically modified food: whether they may provoke an allergic reaction; whether the genes could transfer from the food into human cells; and whether the genes not approved for human consumption could outcross to other crops. There is a scientific consensus that currently available food derived from GM crops poses no greater risk to human health than conventional food, but that each GM food needs to be tested on a case-by-case basis before introduction. Nonetheless, members of the public are less likely than scientists to perceive GM foods as safe.
In popular culture
Genetic engineering features in many science fiction stories. Frank Herbert's novel The White Plague described the deliberate use of genetic engineering to create a pathogen which specifically killed women. Another of Herbert's creations, the Dune series of novels, uses genetic engineering to create the powerful but despised Tleilaxu. Films such as The Island and Blade Runner bring the engineered creature to confront the person who created it or the being it was cloned from. Few films have informed audiences about genetic engineering, with the exception of the 1978 The Boys from Brazil and the 1993 Jurassic Park, both of which made use of a lesson, a demonstration, and a clip of scientific film.
Genetic engineering methods are weakly represented in film; Michael Clark, writing for The Wellcome Trust, calls the portrayal of genetic engineering and biotechnology "seriously distorted" in films such as The 6th Day. In Clark's view, the biotechnology is typically "given fantastic but visually arresting forms" while the science is either relegated to the background or fictionalised to suit a young audience.










![{\displaystyle {\begin{array}{lll}\Delta G_{\text{bind}}=-RT\ln K_{\text{d}}\\[1.3ex]K_{\text{d}}={\dfrac {[{\text{Ligand}}][{\text{Receptor}}]}{[{\text{Complex}}]}}\\[1.3ex]\Delta G_{\text{bind}}=\Delta G_{\text{desolvation}}+\Delta G_{\text{motion}}+\Delta G_{\text{configuration}}+\Delta G_{\text{interaction}}\end{array}}}](https://wikimedia.org/api/rest_v1/media/math/render/svg/ba49ddd9dec7415d129787213744ca1afcd2d021)






