Genome instability (also genetic instability or genomic instability) refers to a high frequency of mutations within the genome of a cellular lineage. These mutations can include changes in nucleic acid sequences, chromosomal rearrangements or aneuploidy. Genome instability does occur in bacteria. In multicellular organisms genome instability is central to carcinogenesis, and in humans it is also a factor in some neurodegenerative diseases such as amyotrophic lateral sclerosis or the neuromuscular disease myotonic dystrophy.
The sources of genome instability have only recently begun to be elucidated. A high frequency of externally caused DNA damage can be one source of genome instability since DNA damage can cause inaccurate translesion DNA synthesis past the damage or errors in repair, leading to mutation. Another source of genome instability may be epigenetic or mutational reductions in expression of DNA repair genes. Because endogenous (metabolically-caused) DNA damage is very frequent, occurring on average more than 60,000 times a day in the genomes of human cells, any reduced DNA repair is likely an important source of genome instability.
Usual genome situation
Usually, all cells in an individual in a given species (plant or animal) show a constant number of chromosomes, which constitute what is known as the karyotype defining this species (see also List of number of chromosomes of various organisms), although some species present a very high karyotypic variability. In humans, mutations that would change an amino acid within the protein coding region of the genome occur at an average of only 0.35 per generation (less than one mutated protein per generation).
Sometimes, in a species with a stable karyotype, random variations that modify the normal number of chromosomes may be observed. In other cases, there are structural alterations (e.g., chromosomal translocations, deletions) that modify the standard chromosomal complement. In these cases, it is indicated that the affected organism presents genome instability (also genetic instability, or even chromosomic instability). The process of genome instability often leads to a situation of aneuploidy, in which the cells present a chromosomic number that is either higher or lower than the normal complement for the species.
Causes of genome instability
DNA Replication Defects
In the cell cycle, DNA is usually most vulnerable during replication. The replisome must be able to navigate obstacles such as tightly wound chromatin with bound proteins, single and double stranded breaks which can lead to the stalling of the replication fork. Each protein or enzyme in the replisome must perform its function well to result in a perfect copy of DNA. Mutations of proteins such as DNA polymerase or DNA ligase can lead to impairment of replication and lead to spontaneous chromosomal exchanges. Proteins such as Tel1 and Mec1 (ATR, ATM in humans) can detect single and double-stranded breaks and recruit factors such as Rmr3 helicase to stabilize the replication fork in order to prevent its collapse. Mutations in Tel1, Mec1, and Rmr3 helicase result in a significant increase of chromosomal recombination. ATR responds specifically to stalled replication forks and single-stranded breaks resulting from UV damage while ATM responds directly to double-stranded breaks. These proteins also prevent progression into mitosis by inhibiting the firing of late replication origins until the DNA breaks are fixed by phosphorylating CHK1 and CHK2, which results in a signaling cascade arresting the cell in S-phase. For single stranded breaks, replication occurs until the location of the break, then the other strand is nicked to form a double stranded break, which can then be repaired by Break Induced Replication or homologous recombination using the sister chromatid as an error-free template. In addition to S-phase checkpoints, G1 and G2 checkpoints exist to check for transient DNA damage which could be caused by mutagens such as UV damage. An example is the Saccharomyces pombe gene rad9 which arrests the cells in late S/G2 phase in the presence of DNA damage caused by radiation. The yeast cells with defective rad9 failed to arrest following irradiation, continued cell division, and died rapidly; the cells with wild-type rad9 successfully arrested in late S/G2 phase and remained viable. The cells that arrested were able to survive due to the increased time in S/G2 phase allowing for DNA repair enzymes to function fully.
Fragile Sites
There are hotspots in the genome where DNA sequences are prone to gaps and breaks after inhibition of DNA synthesis such as in the aforementioned checkpoint arrest. These sites are called fragile sites, and can occur commonly as naturally present in most mammalian genomes or occur rarely as a result of mutations, such as DNA-repeat expansion. Rare fragile sites can lead to genetic disease such as fragile X mental retardation syndrome, myotonic dystrophy, Friedrich's ataxia, and Huntington's disease, most of which are caused by expansion of repeats at the DNA, RNA, or protein level. Although, seemingly harmful, these common fragile sites are conserved all the way to yeast and bacteria. These ubiquitous sites are characterized by trinucleotide repeats, most commonly CGG, CAG, GAA, and GCN. These trinucleotide repeats can form into hairpins, leading to difficulty of replication. Under replication stress, such as defective machinery or further DNA damage, DNA breaks and gaps can form at these fragile sites. Using a sister chromatid as repair is not a fool-proof backup as the surrounding DNA information of the n and n+1 repeat is virtually the same, leading to copy number variation. For example, the 16th copy of CGG might be mapped to the 13th copy of CGG in the sister chromatid since the surrounding DNA is both CGGCGGCGG..., leading to 3 extra copies of CGG in the final DNA sequence.
Transcription-associated instability
In both E. coli and Saccharomyces pombe, transcription sites tend to have higher recombination and mutation rates. The coding or non-transcribed strand accumulates more mutations than the template strand. This is due to the fact that the coding strand is single-stranded during transcription, which is chemically more unstable than double-stranded DNA. During elongation of transcription, supercoiling can occur behind an elongating RNA polymerase, leading to single-stranded breaks. When the coding strand is single-stranded, it can also hybridize with itself, creating DNA secondary structures that can compromise replication. In E. coli, when attempting to transcribe GAA triplets such as those found in Friedrich's ataxia, the resulting RNA and template strand can form mismatched loops between different repeats, leaving the complementary segment in the coding strand available to form its own loops which impede replication. Furthermore, replication of DNA and transcription of DNA are not temporally independent; they can occur at the same time and lead to collisions between the replication fork and RNA polymerase complex. In S. cerevisiae, Rrm3 helicase is found at highly transcribed genes in the yeast genome, which is recruited to stabilize a stalling replication fork as described above. This suggests that transcription is an obstacle to replication, which can lead to increased stress in the chromatin spanning the short distance between the unwound replication fork and transcription start site, potentially causing single-stranded DNA breaks. In yeast, proteins act as barriers at the 3' of the transcription unit to prevent further travel of the DNA replication fork.
Increase Genetic Variability
In some portions of the genome, variability is essential to survival. One such locale is the Ig genes. In a pre-B cell, the region consists of all V, D, and J segments. During development of the B cell, a specific V, D, and J segment is chosen to be spliced together to form the final gene, which is catalyzed by RAG1 and RAG2 recombinases. Activation-Induced Cytidine Deaminase (AID) then converts cytidine into uracil. Uracil normally does not exist in DNA, and thus the base is excised and the nick is converted into a double-stranded break which is repaired by non-homologous end joining (NHEJ). This procedure is very error-prone and leads to somatic hypermutation. This genomic instability is crucial in ensuring mammalian survival against infection. V, D, J recombination can ensure millions of unique B-cell receptors; however, random repair by NHEJ introduces variation which can create a receptor that can bind with higher affinity to antigens.
In neuronal and neuromuscular disease
Of about 200 neurological and neuromuscular disorders, 15 have a clear link to an inherited or acquired defect in one of the DNA repair pathways or excessive genotoxic oxidative stress. Five of them (xeroderma pigmentosum, Cockayne's syndrome, trichothiodystrophy, Down's syndrome, and triple-A syndrome) have a defect in the DNA nucleotide excision repair pathway. Six (spinocerebellar ataxia with axonal neuropathy-1, Huntington's disease, Alzheimer's disease, Parkinson's disease, Down's syndrome and amyotrophic lateral sclerosis) seem to result from increased oxidative stress, and the inability of the base excision repair pathway to handle the damage to DNA that this causes. Four of them (Huntington's disease, various spinocerebellar ataxias, Friedreich's ataxia and myotonic dystrophy types 1 and 2) often have an unusual expansion of repeat sequences in DNA, likely attributable to genome instability. Four (ataxia-telangiectasia, ataxia-telangiectasia-like disorder, Nijmegen breakage syndrome and Alzheimer's disease) are defective in genes involved in repairing DNA double-strand breaks. Overall, it seems that oxidative stress is a major cause of genomic instability in the brain. A particular neurological disease arises when a pathway that normally prevents oxidative stress is deficient, or a DNA repair pathway that normally repairs damage caused by oxidative stress is deficient.
In cancer
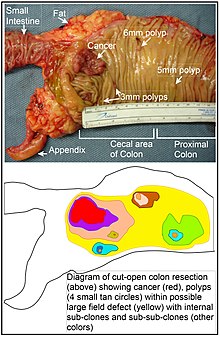
In cancer, genome instability can occur prior to or as a consequence of transformation. Genome instability can refer to the accumulation of extra copies of DNA or chromosomes, chromosomal translocations, chromosomal inversions, chromosome deletions, single-strand breaks in DNA, double-strand breaks in DNA, the intercalation of foreign substances into the DNA double helix, or any abnormal changes in DNA tertiary structure that can cause either the loss of DNA, or the misexpression of genes. Situations of genome instability (as well as aneuploidy) are common in cancer cells, and they are considered a "hallmark" for these cells. The unpredictable nature of these events are also a main contributor to the heterogeneity observed among tumour cells.
It is currently accepted that sporadic tumors (non-familial ones) are originated due to the accumulation of several genetic errors. An average cancer of the breast or colon can have about 60 to 70 protein altering mutations, of which about 3 or 4 may be "driver" mutations, and the remaining ones may be "passenger" mutations Any genetic or epigenetic lesion increasing the mutation rate will have as a consequence an increase in the acquisition of new mutations, increasing then the probability to develop a tumor. During the process of tumorogenesis, it is known that diploid cells acquire mutations in genes responsible for maintaining genome integrity (caretaker genes), as well as in genes that are directly controlling cellular proliferation (gatekeeper genes). Genetic instability can originate due to deficiencies in DNA repair, or due to loss or gain of chromosomes, or due to large scale chromosomal reorganizations. Losing genetic stability will favour tumor development, because it favours the generation of mutants that can be selected by the environment.
The tumor microenvironment has an inhibitory effect on DNA repair pathways contributing to genomic instability, which promotes tumor survival, proliferation, and malignant transformation.
Low frequency of mutations without cancer
The protein coding regions of the human genome, collectively called the exome, constitutes only 1.5% of the total genome. As pointed out above, ordinarily there are only an average of 0.35 mutations in the exome per generation (parent to child) in humans. In the entire genome (including non-protein coding regions) there are only about 70 new mutations per generation in humans.
Cause of mutations in cancer
The likely major underlying cause of mutations in cancer is DNA damage. For example, in the case of lung cancer, DNA damage is caused by agents in exogenous genotoxic tobacco smoke (e.g. acrolein, formaldehyde, acrylonitrile, 1,3-butadiene, acetaldehyde, ethylene oxide and isoprene). Endogenous (metabolically-caused) DNA damage is also very frequent, occurring on average more than 60,000 times a day in the genomes of human cells (see DNA damage (naturally occurring)). Externally and endogenously caused damages may be converted into mutations by inaccurate translesion synthesis or inaccurate DNA repair (e.g. by non-homologous end joining). In addition, DNA damages can also give rise to epigenetic alterations during DNA repair. Both mutations and epigenetic alterations (epimutations) can contribute to progression to cancer.
Very frequent mutations in cancer
As noted above, about 3 or 4 driver mutations and 60 passenger mutations occur in the exome (protein coding region) of a cancer. However, a much larger number of mutations occur in the non-protein-coding regions of DNA. The average number of DNA sequence mutations in the entire genome of a breast cancer tissue sample is about 20,000. In an average melanoma tissue sample (where melanomas have a higher exome mutation frequency) the total number of DNA sequence mutations is about 80,000.
Cause of high frequency of mutations in cancer
The high frequency of mutations in the total genome within cancers suggests that, often, an early carcinogenic alteration may be a deficiency in DNA repair. Mutation rates substantially increase (sometimes by 100-fold) in cells defective in DNA mismatch repair or in homologous recombinational DNA repair. Also, chromosomal rearrangements and aneuploidy increase in humans defective in DNA repair gene BLM.
A deficiency in DNA repair itself can allow DNA damages to accumulate, and error-prone translesion synthesis past some of those damages may give rise to mutations. In addition, faulty repair of these accumulated DNA damages may give rise to epigenetic alterations or epimutations. While a mutation or epimutation in a DNA repair gene itself would not confer a selective advantage, such a repair defect may be carried along as a passenger in a cell when the cell acquires an additional mutation/epimutation that does provide a proliferative advantage. Such cells, with both proliferative advantages and one or more DNA repair defects (causing a very high mutation rate), likely give rise to the 20,000 to 80,000 total genome mutations frequently seen in cancers.
DNA repair deficiency in cancer
In somatic cells, deficiencies in DNA repair sometimes arise by mutations in DNA repair genes, but much more often are due to epigenetic reductions in expression of DNA repair genes. Thus, in a sequence of 113 colorectal cancers, only four had somatic missense mutations in the DNA repair gene MGMT, while the majority of these cancers had reduced MGMT expression due to methylation of the MGMT promoter region. Five reports, listed in the article Epigenetics (see section "DNA repair epigenetics in cancer") presented evidence that between 40% and 90% of colorectal cancers have reduced MGMT expression due to methylation of the MGMT promoter region.
Similarly, for 119 cases of colorectal cancers classified as mismatch repair deficient and lacking DNA repair gene PMS2 expression, Pms2 was deficient in 6 due to mutations in the PMS2 gene, while in 103 cases PMS2 expression was deficient because its pairing partner MLH1 was repressed due to promoter methylation (PMS2 protein is unstable in the absence of MLH1). The other 10 cases of loss of PMS2 expression were likely due to epigenetic overexpression of the microRNA, miR-155, which down-regulates MLH1.
In cancer epigenetics (see section Frequencies of epimutations in DNA repair genes), there is a partial listing of epigenetic deficiencies found in DNA repair genes in sporadic cancers. These include frequencies of between 13–100% of epigenetic defects in genes BRCA1, WRN, FANCB, FANCF, MGMT, MLH1, MSH2, MSH4, ERCC1, XPF, NEIL1 and ATM located in cancers including breast, ovarian, colorectal and head and neck. Two or three epigenetic deficiencies in expression of ERCC1, XPF and/or PMS2 were found to occur simultaneously in the majority of the 49 colon cancers evaluated. Some of these DNA repair deficiencies can be caused by epimutations in microRNAs as summarized in the MicroRNA article section titled miRNA, DNA repair and cancer.