
Ancient DNA (aDNA) is DNA isolated from ancient sources (typically specimens, but also environmental DNA). Due to degradation processes (including cross-linking, deamination and fragmentation) ancient DNA is more degraded in comparison with contemporary genetic material. Genetic material has been recovered from paleo/archaeological and historical skeletal material, mummified tissues, archival collections of non-frozen medical specimens, preserved plant remains, ice and from permafrost cores, marine and lake sediments and excavation dirt.
Even under the best preservation conditions, there is an upper boundary of 0.4–1.5 million years for a sample to contain sufficient DNA for sequencing technologies. The oldest DNA sequenced from physical specimens are from mammoth molars in Siberia over 1 million years old. In 2022, two-million-year-old genetic material was recovered from sediments in Greenland, and is currently considered the oldest DNA discovered so far.
History of ancient DNA studies
1980s

The first study of what would come to be called aDNA was conducted in 1984, when Russ Higuchi and colleagues at the University of California, Berkeley reported that traces of DNA from a museum specimen of the Quagga not only remained in the specimen over 150 years after the death of the individual, but could be extracted and sequenced. Over the next two years, through investigations into natural and artificially mummified specimens, Svante Pääbo confirmed that this phenomenon was not limited to relatively recent museum specimens but could apparently be replicated in a range of mummified human samples that dated as far back as several thousand years.
The laborious processes that were required at that time to sequence such DNA (through bacterial cloning) were an effective brake on the study of ancient DNA (aDNA) and the field of museomics. However, with the development of the Polymerase Chain Reaction (PCR) in the late 1980s, the field began to progress rapidly. Double primer PCR amplification of aDNA (jumping-PCR) can produce highly skewed and non-authentic sequence artifacts. Multiple primer, nested PCR strategy was used to overcome those shortcomings.
1990s

The post-PCR era heralded a wave of publications as numerous research groups claimed success in isolating aDNA. Soon a series of incredible findings had been published, claiming authentic DNA could be extracted from specimens that were millions of years old, into the realms of what Lindahl (1993b) has labelled Antediluvian DNA. The majority of such claims were based on the retrieval of DNA from organisms preserved in amber. Insects such as stingless bees, termites, and wood gnats, as well as plant and bacterial sequences were said to have been extracted from Dominican amber dating to the Oligocene epoch. Still older sources of Lebanese amber-encased weevils, dating to within the Cretaceous epoch, reportedly also yielded authentic DNA. Claims of DNA retrieval were not limited to amber.
Reports of several sediment-preserved plant remains dating to the Miocene were published. Then in 1994, Woodward et al. reported what at the time was called the most exciting results to date — mitochondrial cytochrome b sequences that had apparently been extracted from dinosaur bones dating to more than 80 million years ago. When in 1995 two further studies reported dinosaur DNA sequences extracted from a Cretaceous egg, it seemed that the field would revolutionize knowledge of the Earth's evolutionary past. Even these extraordinary ages were topped by the claimed retrieval of 250-million-year-old halobacterial sequences from halite.
The development of a better understanding of the kinetics of DNA preservation, the risks of sample contamination and other complicating factors led the field to view these results more skeptically. Numerous careful attempts failed to replicate many of the findings, and all of the decade's claims of multi-million year old aDNA would come to be dismissed as inauthentic.
2000s
Single primer extension amplification was introduced in 2007 to address postmortem DNA modification damage. Since 2009 the field of aDNA studies has been revolutionized with the introduction of much cheaper research techniques. The use of high-throughput Next Generation Sequencing (NGS) techniques in the field of ancient DNA research has been essential for reconstructing the genomes of ancient or extinct organisms. A single-stranded DNA (ssDNA) library preparation method has sparked great interest among ancient DNA (aDNA) researchers.

In addition to these technical innovations, the start of the decade saw the field begin to develop better standards and criteria for evaluating DNA results, as well as a better understanding of the potential pitfalls.
2020s
Autumn of 2022, the Nobel Prize of Physiology or Medicine was awarded to Svante Pääbo "for his discoveries concerning the genomes of extinct hominins and human evolution". A few days later, on the 7th of December 2022, a study in Nature reported that two-million year old genetic material was found in Greenland, and is currently considered the oldest DNA discovered so far.
Problems and errors
Degradation processes
Due to degradation processes (including cross-linking, deamination and fragmentation), ancient DNA is of lower quality than modern genetic material. The damage characteristics and ability of aDNA to survive through time restricts possible analyses and places an upper limit on the age of successful samples. There is a theoretical correlation between time and DNA degradation, although differences in environmental conditions complicate matters. Samples subjected to different conditions are unlikely to predictably align to a uniform age-degradation relationship. The environmental effects may even matter after excavation, as DNA decay-rates may increase, particularly under fluctuating storage conditions. Even under the best preservation conditions, there is an upper boundary of 0.4 to 1.5 million years for a sample to contain sufficient DNA for contemporary sequencing technologies.
Research into the decay of mitochondrial and nuclear DNA in moa bones has modelled mitochondrial DNA degradation to an average length of 1 base pair after 6,830,000 years at −5 °C. The decay kinetics have been measured by accelerated aging experiments, further displaying the strong influence of storage temperature and humidity on DNA decay. Nuclear DNA degrades at least twice as fast as mtDNA. Early studies that reported recovery of much older DNA, for example from Cretaceous dinosaur remains, may have stemmed from contamination of the sample.
Age limit
A critical review of ancient DNA literature through the development of the field highlights that few studies have succeeded in amplifying DNA from remains older than several hundred thousand years. A greater appreciation for the risks of environmental contamination and studies on the chemical stability of DNA have raised concerns over previously reported results. The alleged dinosaur DNA was later revealed to be human Y-chromosome. The DNA reported from encapsulated halobacteria has been criticized based on its similarity to modern bacteria, which hints at contamination, or they may be the product of long-term, low-level metabolic activity.
aDNA may contain a large number of postmortem mutations, increasing with time. Some regions of polynucleotide are more susceptible to this degradation, allowing erroneous sequence data to bypass statistical filters used to check the validity of data. Due to sequencing errors, great caution should be applied to interpretation of population size. Substitutions resulting from deamination of cytosine residues are vastly over-represented in the ancient DNA sequences. Miscoding of C to T and G to A accounts for the majority of errors.
Contamination
Another problem with ancient DNA samples is contamination by modern human DNA and by microbial DNA (most of which is also ancient). New methods have emerged in recent years to prevent possible contamination of aDNA samples, including conducting extractions under extreme sterile conditions, using special adapters to identify endogenous molecules of the sample (distinguished from those introduced during analysis), and applying bioinformatics to resulting sequences based on known reads in order to approximate rates of contamination.
Authentication of aDNA
Development in the aDNA field in the 2000s increased the importance of authenticating recovered DNA to confirm that it is indeed ancient and not the result of recent contamination. As DNA degrades over time, the nucleotides that make up the DNA may change, especially at the ends of the DNA molecules. The deamination of cytosine to uracil at the ends of DNA molecules has become a way of authentication. During DNA sequencing, the DNA polymerases will incorporate an adenine (A) across from the uracil (U), leading to cytosine (C) to thymine (T) substitutions in the aDNA data. These substitutions increase in frequency as the sample gets older. Frequency measurement of the C-T level, ancient DNA damage, can be made using various software such as mapDamage2.0 or PMDtools and interactively on metaDMG. Due to hydrolytic depurination, DNA fragments into smaller pieces, leading to single-stranded breaks. Combined with the damage pattern, this short fragment length can also help differentiate between modern and ancient DNA.
Non-human aDNA
Despite the problems associated with aDNA, a wide and ever-increasing range of aDNA sequences have now been published from a range of animal and plant taxa. Tissues examined include artificially or naturally mummified animal remains, bone, shells, paleofaeces, alcohol preserved specimens, rodent middens, dried plant remains, and recently, extractions of animal and plant DNA directly from soil samples.
In June 2013, a group of researchers including Eske Willerslev, Marcus Thomas Pius Gilbert and Orlando Ludovic of the Centre for Geogenetics, Natural History Museum of Denmark at the University of Copenhagen, announced that they had sequenced the DNA of a 560–780 thousand year old horse, using material extracted from a leg bone found buried in permafrost in Canada's Yukon territory. A German team also reported in 2013 the reconstructed mitochondrial genome of a bear, Ursus deningeri, more than 300,000 years old, proving that authentic ancient DNA can be preserved for hundreds of thousand years outside of permafrost. The DNA sequence of even older nuclear DNA was reported in 2021 from the permafrost-preserved teeth of two Siberian mammoths, both over a million years old.
Researchers in 2016 measured chloroplast DNA in marine sediment cores, and found diatom DNA dating back to 1.4 million years. This DNA had a half-life significantly longer than previous research, of up to 15,000 years. Kirkpatrick's team also found that DNA only decayed along a half-life rate until about 100 thousand years, at which point it followed a slower, power-law decay rate.
Human aDNA
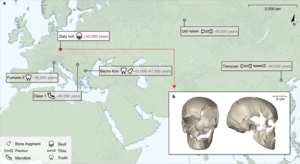
Due to the considerable anthropological, archaeological, and public interest directed toward human remains, they have received considerable attention from the DNA community. There are also more profound contamination issues, since the specimens belong to the same species as the researchers collecting and evaluating the samples.
Sources
Due to the morphological preservation in mummies, many studies from the 1990s and 2000s used mummified tissue as a source of ancient human DNA. Examples include both naturally preserved specimens, such as the Ötzi the Iceman frozen in a glacier and bodies preserved through rapid desiccation at high altitude in the Andes, as well as various chemically treated preserved tissue such as the mummies of ancient Egypt. However, mummified remains are a limited resource. The majority of human aDNA studies have focused on extracting DNA from two sources much more common in the archaeological record: bones and teeth. The bone that is most often used for DNA extraction is the petrous ear bone, since its dense structure provides good conditions for DNA preservation. Several other sources have also yielded DNA, including paleofaeces, and hair. Contamination remains a major problem when working on ancient human material.
Ancient pathogen DNA has been successfully retrieved from samples dating to more than 5,000 years old in humans and as long as 17,000 years ago in other species. In addition to the usual sources of mummified tissue, bones and teeth, such studies have also examined a range of other tissue samples, including calcified pleura, tissue embedded in paraffin, and formalin-fixed tissue. Efficient computational tools have been developed for pathogen and microorganism aDNA analyses in a small (QIIME) and large scale (FALCON).
Results
Taking preventative measures in their procedure against such contamination though, a 2012 study analyzed bone samples of a Neanderthal group in the El Sidrón cave, finding new insights on potential kinship and genetic diversity from the aDNA. In November 2015, scientists reported finding a 110,000-year-old tooth containing DNA from the Denisovan hominin, an extinct species of human in the genus Homo.
The research has added new complexity to the peopling of Eurasia. A study from 2018 showed that a Bronze Age mass migration had greatly impacted the genetic makeup of the British Isles, bringing with it the Bell Beaker culture from mainland Europe.
It has also revealed new information about links between the ancestors of Central Asians and the indigenous peoples of the Americas. In Africa, older DNA degrades quickly due to the warmer tropical climate, although, in September 2017, ancient DNA samples, as old as 8,100 years old, have been reported.
Moreover, ancient DNA has helped researchers to estimate modern human divergence. By sequencing African genomes from three Stone Age hunter gatherers (2000 years old) and four Iron Age farmers (300 to 500 years old), Schlebusch and colleagues were able to push back the date of the earliest divergence between human populations to 350,000 to 260,000 years ago.
As of 2021, the oldest completely reconstructed human genomes are ~45,000 years old. Such genetic data provides insights into the migration and genetic history – e.g. of Europe – including about interbreeding between archaic and modern humans like a common admixture between initial European modern humans and Neanderthals.











