
Knotted proteins are proteins whose backbones entangle themselves in a knot. One can imagine pulling a protein chain from both termini, as though pulling a string from both ends. When a knotted protein is “pulled” from both termini, it does not get disentangled. Knotted proteins are very rare, making up only about one percent of the proteins in the Protein Data Bank, and their folding mechanisms and function are not well understood. Although there are experimental and theoretical studies that hint to some answers, systematic answers to these questions have not yet been found.
Although number of computational methods have been developed to detect protein knots, there are still no completely automatic methods to detect protein knots without necessary manual intervention due to the missing residues or chain breaks in the X-ray structures or the nonstandard PDB formats.
Most of the knots discovered in proteins are deep trefoil (31) knots. Figure eight knots (41), three-twist knots (52), Stevedore knots (61) and Septoil knot (71) have also been discovered. Recently, use of machine learning techniques for predicting protein structure, resulted in highly accurate prediction of 63 knot. Furthermore, using same techniques, composite knots (namely 31#31) were found and crystallised.

Mathematical interpretation
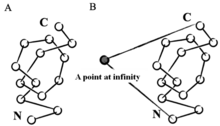
Mathematically, a knot is defined as a subset of three-dimensional space that is homeomorphic to a circle. According to this definition, a knot must exist in a closed loop, while knotted proteins instead exist within open, unclosed chains. In order to apply mathematical knot theory to knotted proteins, various strategies can be used to create an artificial closed loop. One such strategy is to choose a point in space at infinite distance to be connected to the protein's N- and C-termini through a virtual bond, thus the protein can be treated as a closed loop. Another such strategy is to use stochastic methods that create random closures.
Depth of the knot
The depth of a protein knot relates to the ability of the protein to resist unknotting. A deep knot is preserved even though the removal of a considerable number of residues from either end does not destroy the knot. The higher the number of residues that can be removed without destroying the knot, the deeper the knot.
Formation of knots
Considering how knots may be produced with a string, the folding of knotted proteins should involve first the formation of a loop, and then the threading of one terminus through the loop. This is the only topological way that the trefoil knot can be formed. For more complex knots, it is theoretically possible to have the loop to twist multiple times around itself, meaning that one end of the chain gets wrapped around at least once, and then threading to occur. It has also been observed in a theoretical study that a 61 knot can form by the C-terminus threading through a loop, and another loop flipping over the first loop, as well as the C-terminus threading through both the loops which have previously flipped over each other.
The folding of knotted proteins may be explained by interaction of the nascent chain with the ribosome. In particular, the affinity of the chain to the ribosome surface may result in creation of the loop, which may be next threaded by a nascent chain. Such mechanism was shown to be plausible for one of the most deeply knotted proteins known.
There have been experimental studies involving YibK and YbeA, knotted proteins containing trefoil knots. It has been established that these knotted proteins fold slowly, and that the knotting in folding is the rate limiting step. In another experimental study, a 91-residue-long protein was attached to the termini of YibK and YbeA. Attaching the protein to both termini produces a deep knot with about 125 removable residues on each terminus before the knot is destroyed. Yet it was seen that the resulting proteins could fold spontaneously. The attached proteins were shown to fold more quickly than YibK and YbeA themselves, so during folding they are expected to act as plugs at either end of YibK and YbeA. It was found that attaching the protein to the N-terminus did not alter the folding speed, but the attachment to the C-terminus slows folding down, suggesting that the threading event happens at the C-terminus. The chaperones, although facilitate the protein knotting, are not crucial in proteins' self-tying.
Other topologically complex structures in proteins

The class of knotted proteins contains only structures, for which the backbone, after closure forms a knotted loop. However, some proteins contain "internal knots" called slipknots, i.e. unknotted structures containing a knotted subchain. Another topologically complex structure is the link formed by covalent loops, closed by disulfide bridges. Three types of disulfide-based links were identified in proteins: two versions of Hopf link (differing in chirality) and one version of Solomon link. Another complex structure arising by closing part of the chain with covalent bridge are complex lasso proteins, for which the covalent loop is threaded by the chain one or more times. Yet another complex structures arising as a result of the existence of disulfide bridges are the cystine knots, for which two disulfide bridges form a closed, covalent loop, which is threaded by third chain. The term "knot" in the name of the motif is misleading, as the motif does not contain any knotted closed cycle. Moreover, formation of the cystine knots in general is not different from the folding of an unknotted protein
Apart from closing only one chain, one may perform also the chain closure procedure for all the chains present in the crystal structure. In some cases one obtains the non-trivially linked structures, called probabilistic links.
One can also consider loops in proteins formed by pieces of the main chain and the disulfide bridges and interaction via ions. Such loops can also be knotted of form links even within structures with unknotted main chain.
First discoveries
Marc L. Mansfield proposed in 1994, that there can be knots in proteins. He gave unknot scores to proteins by constructing a sphere centered at the center of mass of the alpha carbons of the backbone, with a radius twice the distance between the center of mass and the Calpha that is the farthest away from the center of mass, and by sampling two random points on the surface of the sphere. He connected the two points by tracing a geodesic on the surface of the sphere (arcs of great circles), and then connected each end of the protein chain with one of these points. Repeating this procedure a 100 times and counting the times where the knot is destroyed in the mathematical sense yields the unknot score. Human carbonic anhydrase was identified to have a low unknot score (22). Upon visually inspecting the structure, it was seen that the knot was shallow, meaning that the removal of a few residues from either end destroys the knot.
In 2000, William R. Taylor identified a deep knot in acetohydroxy acid isomeroreductase (PDB ID: 1YVE), by using an algorithm that smooths protein chains and makes knots more visible. The algorithm keeps both termini fixed, and iteratively assigns to the coordinates of each residue the average of the coordinates of the neighboring residues. It has to be made sure that the chains do not pass through each other, otherwise the crossings and therefore the knot might get destroyed. If there is no knot, the algorithm eventually produces a straight line that joins both termini.
Studies about the function of the knot in a protein
Some proposals about the function of knots have been that it might increase thermal and kinetic stability. One particular suggestion was that for the human ubiquitin hydrolase, which contains a 52 knot, the presence of the knot might be preventing it from being pulled into the proteasome. Because it is a deubiquitinating enzyme, it is often found in proximity of proteins soon to be degraded by proteasome, and therefore it faces the danger of being degraded itself. Therefore, the presence of the knot might be functioning as a plug that prevents it. This notion was further analyzed on other proteins like YbeA and YibK with computer simulations. The knots seem to tighten when they are pulled into a pore, and depending on the force with which they are pulled in, they either get stuck and block the pore, the likeliness of which increases with stronger pulling forces, or in the case of a small pulling force they might get disentangled as one terminus is pulled out of the knot. For deeper knots, it is more likely that the pore will be blocked, as there are too many residues that need to be pulled through the knot. In another theoretical study, it was found that the modeled knotted protein was not thermally stable, but it was kinetically stable. It was also shown that the knot in proteins creates the places on the verge of hydrophobic and hydrophilic parts of the chain, characteristic for active sites. This may explain why over 80% of knotted proteins are enzymes. Another study shows that knotted and slipknotted proteins constitute a significant number of membrane proteins. They comprise one of the largest groups of secondary active transporters.
Web servers to extrapolate knotted proteins
Some local programs and a number of web servers are available, providing convenient query services for knotted structures and analysis tools for detecting protein knots, including:
- Topoly - Python package to analyze topology of polymers
- Knot_pull - Python package for biopolymer smoothing and knot detection
- KnotProt 2.0 - Database of proteins with knots and other entangled structures
- AlphaKnot 2.0 - Database and server to analyze entanglement in structures predicted by AlphaFold methods
- pKNOT - Web server for knot detection in proteins