Nucleic acid structure refers to the structure of nucleic acids such as DNA and RNA. Chemically speaking, DNA and RNA are very similar. Nucleic acid structure is often divided into four different levels: primary, secondary, tertiary and quaternary.
Primary structure
Chemical structure of DNA
Primary structure consists of a linear sequence of nucleotides that are linked together by phosphodiester bonds. It is this linear sequence of nucleotides that make up the Primary structure of DNA or RNA. Nucleotides consist of 3 components:
- Nitrogenous base
- 5-carbon sugar which is called deoxyribose (found in DNA) and ribose (found in RNA).
- One or more phosphate groups.[1]
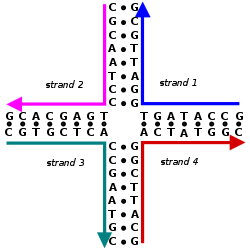
Nucleic acid design can be used to create nucleic acid complexes with complicated secondary structures such as this four-arm junction. These four strands associate into this structure because it maximizes the number of correct base pairs, with A's matched to T's and C's matched to G's. Image from Mao, 2004.[5]
Complexes with alkali metal ions
There are three potential metal binding groups on nucleic acids: phosphate, sugar and base moieties. Solid-state structure of complexes with alkali metal ions have been reviewed.[6]Secondary structure
Secondary structure is the set of interactions between bases, i.e., which parts of strands are bound to each other. In DNA double helix, the two strands of DNA are held together by hydrogen bonds. The nucleotides on one strand base pairs with the nucleotide on the other strand. The secondary structure is responsible for the shape that the nucleic acid assumes. The bases in the DNA are classified as purines and pyrimidines. The purines are adenine and guanine. Purines consist of a double ring structure, a six membered and a five membered ring containing nitrogen. The pyrimidines are cytosine and thymine. It has a single ringed structure, a six membered ring containing nitrogen. A purine base always pairs with a pyrimidine base (guanine (G) pairs with cytosine (C) and adenine (A) pairs with thymine (T) or uracil (U)). DNA's secondary structure is predominantly determined by base-pairing of the two polynucleotide strands wrapped around each other to form a double helix. Although the two strands are aligned by hydrogen bonds in base pairs, the stronger forces holding the two strands together are stacking interactions between the bases. These stacking interactions are stabilized by Van der Waals forces and hydrophobic interactions, and show a large amount of local structural variability.[7] There are also two grooves in the double helix, which are called major groove and minor groove based on their relative size.The secondary structure of RNA consists of a single polynucleotide. Base pairing in RNA occurs when RNA folds between complementarity regions. Both single- and double-stranded regions are often found in RNA molecules. The antiparallel strands form a helical shape.[3] The four basic elements in the secondary structure of RNA are helices, loops, bulges, and junctions. Stem-loop or hairpin loop is the most common element of RNA secondary structure.[8] Stem-loop is formed when the RNA chains fold back on themselves to form a double helical tract called the stem, the unpaired nucleotides forms single stranded region called the loop.[9] Secondary structure of RNA can be predicted by experimental data on the secondary structure elements, helices, loops and bulges. Bulges and internal loops are formed by separation of the double helical tract on either one strand (bulge) or on both strands (internal loops) by unpaired nucleotides. A tetraloop is a four-base pairs hairpin RNA structure. There are three common families of tetraloop in ribosomal RNA: UNCG, GNRA, and CUUG (N is one of the four nucleotides and R is a purine).UNCG is the most stable tetraloop.[10] Pseudoknot is a RNA secondary structure first identified in turnip yellow mosaic virus.[11] Pseudoknots are formed when nucleotides from the hairpin loop pairs with a single stranded region outside of the hairpin to form a helical segment. H-type fold pseudoknots are best characterized. In H-type fold, nucleotides in the hairpin loop pairs with the bases outside the hairpin stem forming second stem and loop. This causes formation of pseudoknots with two stems and two loops.[12] Pseudoknots are functional elements in RNA structure having diverse function and found in most classes of RNA. DotKnot-PW method is used for comparative pseudoknots prediction. The main points in the DotKnot-PW method is scoring the similarities found in stems, secondary elements and H-type pseudoknots.[13]
Tertiary structure
DNA structure and bases
A-B-Z-DNA Side View
Tertiary structure refers to the locations of the atoms in three-dimensional space, taking into consideration geometrical and steric constraints. It is a higher order than the secondary structure, in which large-scale folding in a linear polymer occurs and the entire chain is folded into a specific 3-dimensional shape. There are 4 areas in which the structural forms of DNA can differ.
- Handedness – right or left
- Length of the helix turn
- Number of base pairs per turn
- Difference in size between the major and minor grooves[3]
B-DNA is the most common form of DNA in vivo and is a more narrow, elongated helix than A-DNA. Its wide major groove makes it more accessible to proteins. On the other hand, it has a narrow minor groove. B-DNA's favored conformations occur at high water concentrations; the hydration of the minor groove appears to favor B-DNA. B-DNA base pairs are nearly perpendicular to the helix axis. The sugar pucker which determines the shape of the a-helix, whether the helix will exist in the A-form or in the B-form, occurs at the C2'-endo.[14]
A-DNA, is a form of the DNA duplex observed under dehydrating conditions. It is shorter and wider than B-DNA. RNA adopts this double helical form, and RNA-DNA duplexes are mostly A-form, but B-form RNA-DNA duplexes have been observed.[15] In localized single strand dinucleotide contexts, RNA can also adopt the B-form without pairing to DNA.[16] A-DNA has a deep, narrow major groove which does not make it easily accessible to proteins. On the other hand, its wide, shallow minor groove makes it accessible to proteins but with lower information content than the major groove. Its favored conformation is at low water concentrations. A-DNAs base pairs are tilted relative to the helix axis, and are displaced from the axis. The sugar pucker occurs at the C3'-endo and in RNA 2'-OH inhibits C2'-endo conformation.[14] Long considered little more than a laboratory artifice, A-DNA is now known to have several biological functions.
Z-DNA is a relatively rare left-handed double-helix. Given the proper sequence and superhelical tension, it can be formed in vivo but its function is unclear. It has a more narrow, more elongated helix than A or B. Z-DNA's major groove is not really a groove, and it has a narrow minor groove. The most favored conformation occurs when there are high salt concentrations. There are some base substitutions but they require an alternating purine-pyrimidine sequence. The N2-amino of G H-bonds to 5' PO, which explains the slow exchange of protons and the need for the G purine. Z-DNA base pairs are nearly perpendicular to the helix axis. Z-DNA does not contain single base-pairs but rather a GpC repeat with P-P distances varying for GpC and CpG. On the GpC stack there is good base overlap, whereas on the CpG stack there is less overlap. Z-DNA's zigzag backbone is due to the C sugar conformation compensating for G glycosidic bond conformation. The conformation of G is syn, C2'-endo and for C it is anti, C3'-endo.[14]
A linear DNA molecule having free ends can rotate, to adjust to changes of various dynamic processes in the cell, by changing how many times the two chains of its double helix twist around each other. Some DNA molecules are circular and are topologically constrained. More recently circular RNA was described as well to be a natural pervasive class of nucleic acids, expressed in many organisms (see circRNA).
A covalently closed, circular DNA (also known as cccDNA) is topologically constrained as the number of times the chains coiled around one other cannot change. This cccDNA can be supercoiled, which is the tertiary structure of DNA. Supercoiling is characterized by the linking number, twist and writhe. The linking number (Lk) for circular DNA is defined as the number of times one strand would have to pass through the other strand to completely separate the two strands. The linking number for circular DNA can only be changed by breaking of a covalent bond in one of the two strands. Always an integer, the linking number of a cccDNA is the sum of two components: twists (Tw) and writhes (Wr).[17]