Group I: dsDNA viruses
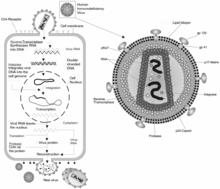
Genome of human herpesvirus-6, a member of the family Herpesviridae
Genome organization within this group varies considerably. Some have circular genomes (Baculoviridae, Papovaviridae and Polydnaviridae) while others have linear genomes (Adenoviridae, Herpesviridae and some phages). Some families have circularly permuted linear genomes (phage T4 and some Iridoviridae). Others have linear genomes with covalently closed ends (Poxviridae and Phycodnaviridae).
A virus infecting archaea
was first described in 1974. Several others have been described since:
most have head-tail morphologies and linear double-stranded DNA genomes.
Other morphologies have also been described: spindle shaped, rod
shaped, filamentous, icosahedral and spherical. Additional morphological
types may exist.
Orders
within this group are defined on the basis of morphology rather than
DNA sequence similarity. It is thought that morphology is more conserved
in this group than sequence similarity or gene order which is extremely
variable. Three orders and 31 families are currently recognised. A fourth order — Megavirales — for the nucleocytoplasmic large DNA viruses has been proposed. This proposal has yet to be ratified by the ICTV. Four genera are recognised that have not yet been assigned a family.
Fifteen families are enveloped. These include all three families in the order Herpesvirales and the following families: Ascoviridae, Ampullaviridae, Asfarviridae, Baculoviridae, Fuselloviridae, Globuloviridae, Guttaviridae, Hytrosaviridae, Iridoviridae, Lipothrixviridae, Nimaviridae and Poxviridae.
Bacteriophages (viruses infecting bacteria) belonging to the families Tectiviridae and Corticoviridae have a lipid bilayer membrane inside the icosahedral protein capsid and the membrane surrounds the genome. The crenarchaeal virus Sulfolobus turreted icosahedral virus has a similar structure.
The genomes in this group vary considerably from ~10 kilobases to
over 2.5 megabases in length. The largest bacteriophage known is
Klebsiella Phage vB_KleM-RaK2 which has a genome of 346 kilobases.
The virophages are a group of viruses that infect other viruses.
A virus with a novel method of genome packing infecting species of the genus Sulfolobus has been described. As this virus does not resemble any known virus it has been classified into a new family, the Portogloboviridae.
Another Sulfolobus infecting virus - Sulfolobus ellipsoid virus 1 - has been described.[5] This enveloped virus has a unique capsid and may be classified into a new taxon.
Host range
Species of the order Caudovirales and of the families Corticoviridae and Tectiviridae infect bacteria.
Species of the order Ligamenvirales and the families Ampullaviridae, Bicaudaviridae, Clavaviridae, Fuselloviridae, Globuloviridae, Guttaviridae and Turriviridae infect hyperthermophilic archaea species of the Crenarchaeota.
Species of the order Herpesvirales and of the families Adenoviridae, Asfarviridae, Iridoviridae, Papillomaviridae, Polyomaviridae and Poxviridae infect vertebrates.
Species of the families Ascovirus, Baculovirus, Hytrosaviridae, Iridoviridae and Polydnaviruses and of the genus Nudivirus infect insects.
Species of the family Mimiviridae and the species Marseillevirus, Megavirus, Mavirus virophage and Sputnik virophage infect protozoa.
Species of the family Nimaviridae infect crustaceans.
Species of the family Phycodnaviridae and the species Organic Lake virophage infect algae. These are the only known dsDNA viruses that infect plants.
Species of the family Plasmaviridae infect species of the class Mollicutes.
Species of the family Pandoraviridae infect amoebae.
Species of the genus Dinodnavirus infect dinoflagellates. These are the only known viruses that infect dinoflagellates.
Species of the genus Rhizidiovirus infect stramenopiles. These are the only known dsDNA viruses that infect stramenopiles.
Species of the genus Salterprovirus and Sphaerolipoviridae infect species of the Euryarchaeota.
Taxonomy
- Order Caudovirales
- Family Myoviridae—includes Enterobacteria phage T4
- Family Podoviridae—includes Enterobacteria phage T7
- Family Siphoviridae—includes Enterobacteria phage λ
- Order Herpesvirales
- Family Alloherpesviridae
- Family Herpesviridae—includes human herpesviruses, Varicella Zoster virus
- Family Malacoherpesviridae
- Order Ligamenvirales
- Family Lipothrixviridae
- Family Rudiviridae
- Unassigned families
- Family Adenoviridae—includes viruses which cause human adenovirus infection
- Family Ampullaviridae
- Family Ascoviridae
- Family Asfarviridae—includes African swine fever virus
- Family Baculoviridae
- Family Bicaudaviridae
- Family Clavaviridae
- Family Corticoviridae
- Family Fuselloviridae
- Family Globuloviridae
- Family Guttaviridae
- Family Hytrosaviridae
- Family Iridoviridae
- Family Lavidaviridae
- Family Marseilleviridae
- Family Mimiviridae
- Family Nudiviridae
- Family Nimaviridae
- Family Pandoraviridae
- Family Papillomaviridae
- Family Phycodnaviridae
- Family Plasmaviridae
- Family Polydnaviruses
- Family Polyomaviridae—includes Simian virus 40, JC virus, BK virus
- Family Poxviridae—includes Cowpox virus, smallpox
- Family Sphaerolipoviridae
- Family Tectiviridae
- Family Turriviridae
- Unassigned genera
- Unassigned species
- Abalone shriveling syndrome-associated virus
- Bandicoot papillomatosis carcinomatosis virus
- Cedratvirus
- Kaumoebavirus
- KIs-V
- Lentille virus
- Leptopilina boulardi filamentous virus
- Megavirus
- Metallosphaera turreted icosahedral virus
- MetSV
- Methanosarcina spherical virus
- Mollivirus sibericum virus
- Orpheovirus IHUMI-LCC2
- Phaeocystis globosa virus
- Pithovirus
- Virophages
- Family Lavidaviridae
- Ace Lake Mavirus virophage
- Dishui Lake virophage 1
- Phaeocystis globosa virus virophage
- Rio Negro virophage
- Sputnik virophage 2
- Yellowstone Lake virophage 1
- Yellowstone Lake virophage 2
- Yellowstone Lake virophage 3
- Yellowstone Lake virophage 4
- Yellowstone Lake virophage 5
- Yellowstone Lake virophage 6
- Yellowstone Lake virophage 7
- Zamilon virophage 2
Unclassified viruses
A group of double stranded DNA viruses have been found in fish that appear to be related to the herpesviruses.
Another group of viruses that infect fish has been described.
Pleolipoviruses
A
group known as the pleolipoviruses, although having a similar genome
organisation, differ in having either single or double stranded DNA
genomes. Within the double stranded forms have runs of single stranded DNA. These viruses have been placed in the family Pleolipoviridae. This family has been divided in three genera: Alphapleolipovirus, Betapleolipovirus and Gammapleolipovirus.
These viruses are nonlytic and form virions characterized by a lipid vesicle enclosing the genome.
They do not have nucleoproteins. The lipids in the viral membrane are
unselectively acquired from host cell membranes. The virions contain two
to three major structural proteins, which either are embedded in the membrane or form spikes distributed randomly on the external membrane surface.
This group includes the following viruses:
- Genus: Alphapleolipovirus
- Haloarcula hispanica pleomorphic virus 1 (Haloarcula virus HHPV1)
- Haloarcula hispanica pleomorphic virus 2 (Haloarcula virus HHPV2)
- Halorubrum pleomorphic virus 1
- Halorubrum pleomorphic virus 2
- Halorubrum pleomorphic virus 6
- Genus: Betapleolipovirus
- Halogeometricum pleomorphic virus 1 (Halogeometricum virus HGPV1)
- Halorubrum pleomorphic virus 3 (Halorubrum virus HRPV3)
- SNJ2
- Genus: Gammapleolipovirus
Group II: ssDNA viruses
Genome of bacteriophage ΦX174, a single-stranded DNA virus
Although bacteriophages were first described in 1927, it was only in 1959 that Sinshemer working with phage Phi X 174 showed that they could possess single-stranded DNA genomes. Despite this discovery, until relatively recently it was believed that most DNA viruses contained double-stranded DNA. Recent work, however, has shown that single-stranded DNA viruses can be highly abundant in seawater, freshwater, sediments, terrestrial and extreme environments, as well as metazoan-associated and marine microbial mats. Many of these "environmental" viruses belong to the family Microviridae.
However, the vast majority has yet to be classified and assigned to
genera and higher taxa. Because most of these viruses do not appear to
be related, or are only distantly related to known viruses, additional
taxa will have to be created to accommodate them.
- Archaea
Although ~50 archaeal viruses are known, all but two have double
stranded genomes. These two viruses have been placed in the families Pleolipoviridae and Spiraviridae
Taxonomy
Families
in this group have been assigned on the basis of the nature of the
genome (circular or linear) and the host range. Eleven families are
currently recognised.
- Family Anelloviridae
- Family Bacilladnavirus
- Family Bidnaviridae
- Family Circoviridae
- Family Geminiviridae
- Family Genomoviridae
- Family Inoviridae
- Family Microviridae
- Family Nanoviridae
- Family Parvoviridae
- Family Smacoviridae
- Family Spiraviridae
Classification
A division of the circular single stranded viruses into four types has been proposed. This division seems likely to reflect their phylogenetic relationships.
Type I genomes are characterized by a small circular DNA genome
(approximately 2-kb), with the Rep protein and the major open reading
frame (ORF) in opposite orientations. This type is characteristic of the
circoviruses, geminiviruses and nanoviruses.
Type II genomes have the unique feature of two separate Rep ORFs.
Type III genomes contain two major ORFs in the same orientation. This arrangement is typical of the anelloviruses.
Type IV genomes have the largest genomes of nearly 4-kb, with up
to eight ORFs. This type of genome is found in the Inoviridae and the
Microviridae.
Given the variety of single stranded viruses that have been
described this scheme—if it is accepted by the ICTV—will need to be
extended.
CRESS viruses
All
known eukaryotic ssDNA viruses also form icosahedral capsids. With the
exception of the family Bidnaviridae, all eukaryotic ssDNA viruses
encode homologous rolling-circle replication initiation proteins with
characteristic N-terminal endonuclease domains and C-terminal
superfamily three helicase domains. A name for this group of viruses has been proposed - circular Rep-encoding single-strand (CRESS) DNA viruses.
Cruciviridae
A
group of ssDNA viruses whose Rep proteins show homology to ssDNA
viruses from the families Geminiviridae, Circoviridae, and Nanoviridae,
while their coat protein is related to those of ssRNA viruses from the
family Tombusviridae and unclassified oomycete-infecting viruses. The name Cruciviridae has been proposed for this group.
Host range
The families Bidnaviridae and Parvoviridae have linear genomes while the other families have circular genomes. The Bidnaviridae have a two part genome and infect invertebrates. The Inoviridae and Microviridae infect bacteria; the Anelloviridae and Circoviridae infect animals (mammals and birds respectively); and the Geminiviridae and Nanoviridae infect plants. In both the Geminiviridae and Nanoviridae the genome is composed of more than a single chromosome. The Bacillariodnaviridae infect diatoms
and have a unique genome: the major chromosome is circular (~6
kilobases in length): the minor chromosome is linear (~1 kilobase in
length) and complementary to part of the major chromosome. Members of
the Spiraviridae infect archaea. Members of the Genomoviridae infect fungi.
Molecular biology
All
viruses in this group require formation of a replicative form—a double
stranded DNA intermediate—for genome replication. This is normally
created from the viral DNA with the assistance of the host's own DNA polymerase.
Recently classified viruses
In
the 9th edition of the viral taxonomy of the ICTV (published 2011) the
Bombyx mori densovirus type 2 was placed in a new family—the Bidnaviridae
on the basis of its genome structure and replication mechanism. This is
currently the only member of this family but it seems likely that other
species will be allocated to this family in the near future.
A new genus — Bufavirus — was proposed on the basis of the isolation of two new viruses from human stool. Another member of this genus—megabat bufavius 1—has been reported from bats. The human viruses have since been renamed Primate protoparvovirus and been placed in the genus Protoparvovirus.
Another proposed genus is Pecovirus. These are similar in organisation to the Smacovirus but share little sequence similarity.
The most recently introduced family of ssDNA viruses is the Genomoviridae (the family name is an acronym derived from geminivirus-like, no movement protein).
The family includes 9 genera, namely Gemycircularvirus, Gemyduguivirus, Gemygorvirus, Gemykibivirus, Gemykolovirus, Gemykrogvirus, Gemykroznavirus, Gemytondvirus and Gemyvongvirus.
The genus name Gemycircularvirus stands for Gemini-like myco-infecting circular virus. the type species of the genus Gemycircularvirus - Sclerotinia sclerotiorum hypovirulence associated DNA virus 1 - is currently the only cultivated member of the family. The rest of genomoviruses are uncultivated and have been discovered using metagenomics techniques.
Another genus has been proposed - Gemybolavirus.
- Human isolates
Isolates from this group have also been isolated from the cerebrospinal fluid and brains of patients with multiple sclerosis.
A isolate from this group has also been identified in a child with encephalitis.
Viruses from this group have also been isolated from the blood of HIV+ve patients.
- Animal isolates
Ostrich faecal associated ssDNA virus has been placed in the genus Gemytondvirus. Rabbit faecal associated ssDNA virus has been placed in the genus Gemykroznavirus.
Another virus from this group has been isolated from mosquitoes.
Ten new circular viruses have been isolated from dragonfly larvae.
The genomes range from 1628 to 2668 nucleotides in length. These
dragonfly viruses have since been placed in the Gemycircularviridae.
Additional viruses from this group have been reported from dragonflies and damselflies.
- Plants and fungi
Three viruses in this group have been isolated from plants.
A virus — Cassava associated circular DNA virus — that has some similarity to Sclerotinia sclerotiorum hypovirulence associated DNA virus 1 has been isolated. This virus has been placed in the Gemycircularviridae.
Some of this group of viruses may infect fungi.
Smacoviridae
A new family, the Smacoviridae, has been created for a number of single-stranded DNA viruses isolated from the faeces of various mammals.
Smacoviruses have circular genomes of ~2.5 kilobases and have a Rep
protein and capsid protein encoded in opposite orientations. 43 species
have been included in this family which includes six genera - Bovismacovirus, Cosmacovirus, Dragsmacovirus, Drosmacovirus, Huchismacovirus and Porprismacovirus.
Unassigned species
A number of additional single stranded DNA viruses have been described but are as yet unclassified.
Human isolates
Viruses in this group have been isolated from other cases of encephalitis, diarrhoea and sewage.
Two viruses have been isolated from human feces — circo-like
virus Brazil hs1 and hs2 — with genome lengths of 2526 and 2533
nucleotides respectively.
These viruses have four open reading frames. These viruses appear to be
related to three viruses previously isolated from waste water, a bat
and from a rodent.
This appears to belong to a novel group.
A novel species of virus - human respiratory-associated PSCV-5-like virus - has been isolated from the respiratory tract.
The virus is approximately 3 kilobases in length and has two open
reading frames - one encoding the coat protein and the other the DNA
replicase. The significance - if any - of this virus for human disease
is unknown presently.
Animal viruses — vertebrates
An unrelated group of ssDNA viruses, also discovered using viral metagenomics, includes the species bovine stool associated circular virus and chimpanzee stool associated circular virus. The closest relations to this genus appear to be the Nanoviridae
but further work will be needed to confirm this. Another isolate that
appears to be related to these viruses has been isolated from pig faeces
in New Zealand. This isolate also appears to be related to the pig stool-associated single-stranded DNA virus.
This virus has two large open reading frames one encoding the capsid
gene and the other the Rep gene. These are bidirectionally transcribed
and separated by intergenic regions. Another virus of this group has
been reported again from pigs. A virus from this group has been isolated from turkey faeces. Another ten viruses from this group have been isolated from pig faeces.
Viruses that appear to belong to this group have been isolated from
other mammals including cows, rodents, bats, badgers and foxes.
Another virus in this group has been isolated from birds.
Fur seal feces-associated circular DNA virus was isolated from the feces of a fur seal (Arctocephalus forsteri) in New Zealand. The genome has 2 main open reading frames and is 2925 nucleotides in length. Another virus - porcine stool associated virus 4 - has been isolated. It appears to be related to the fur seal virus.
Two viruses have been described from the nesting material yellow crowned parakeet (Cyanoramphus auriceps) — Cyanoramphus nest-associated circular X virus (2308 nt) and Cyanoramphus nest-associated circular K virus (2087 nt)
Both viruses have two bidirectional open reading frames. Within these
are the rolling-circle replication motifs I, II, III and the helicase
motifs Walker A and Walker B. There is also a conserved nonanucleotide
motif required for rolling-circle replication. CynNCKV has some
similarity to the picobiliphyte nano-like virus (Picobiliphyte M5584-5) and CynNCXV has some similarity to the rodent stool associated virus (RodSCV M-45).
A virus with a circular genome — sea turtle tornovirus 1 — has been isolated from a sea turtle with fibropapillomatosis. It is sufficiently unrelated to any other known virus that it may belong to a new family. The closest relations seem to be the Gyrovirinae. The proposed genus name for this virus is Tornovirus.
Another fecal virus - feline stool-associated circular DNA virus - has been described.
Animal viruses — invertebrates
Among
these are the parvovirus-like viruses. These have linear
single-stranded DNA genomes but unlike the parvoviruses the genome is
bipartate. This group includes Hepatopancreatic parvo-like virus and Lymphoidal parvo-like virus. A new family Bidensoviridae has been proposed for this group but this proposal has not been ratified by the ICTV to date. Their closest relations appear to be the Brevidensoviruses (family Parvoviridae).
A virus — Acheta domesticus volvovirus - has been isolated from the house cricket (Acheta domesticus).
The genome is circular, has four open reading frames and is 2,517
nucleotides in length. It appears to be unrelated to previously
described species. The genus name Volvovirus has been proposed for these species. The genomes in this genus are ~2.5 nucleotides in length and encode 4 open reading frames.
Two new viruses have been isolated from the copepods Acartia tonsa and Labidocera aestiva— Acartia tonsa copepod circo-like virus and Labidocera aestiva copepod circo-like virus respectively.
A virus has been isolated from the mud flat snail (Amphibola crenata).
This virus has a single stranded circular genome of 2351 nucleotides
that encoded 2 open reading frames that are oriented in opposite
directions. The smaller open reading frame (874 nucleotides) encodes a
protein with similarities to the Rep (replication) proteins of circoviruses and plasmids. The larger open reading frame (955 nucleotides) has no homology to any currently known protein.
An unusual — and as yet unnamed — virus has been isolated from the flatworm Girardia tigrina.
Because of its genome organisation, this virus appears to belong to an
entirely new family. It is the first virus to be isolated from a flatworm.
From the hepatopancreas of the shrimp (Farfantepenaeus duorarum) a circular single stranded DNA virus has been isolated. This virus does not appear to cause disease in the shrimp.
A circo-like virus has been isolated from the shrimp (Penaeus monodon). The 1,777-nucleotide genome is circular and single stranded. It has some similarity to the circoviruses and cycloviruses.
Ten viruses have been isolated from echinoderms. All appear to belong to as yet undescribed genera.
Plants
A circular single stranded DNA virus has been isolated from a grapevine.
This species may be related to the family Geminiviridae but differs
from this family in a number of important respects including genome
size.
Several viruses — baminivirus, nepavirus and niminivirus — related to geminvirus have also been reported.
A virus - Ancient caribou feces associated virus - has been cloned from 700-y-old caribou feces.
A new virus with a three part single stranded genome has been reported. This seems likely to be a member of a new family of viruses.
Marine and other
More than 600 single-stranded DNA viral genomes were identified in ssDNA purified from seawater .
These fell into 129 genetically distinct groups that had no
recognizable similarity to each other or to other virus sequences, and
thus many likely represent new families of viruses. Of the 129 groups,
eleven were much more abundant than the others, and although their hosts
have yet to be identified, they are likely to be eukaryotic
phytoplankton, zooplankton and bacteria.
A virus — Boiling Springs Lake virus — appears to have evolved by a recombination event between a DNA virus (circovirus) and an RNA virus (tombusvirus). The genome is circular and encodes two proteins—a Rep protein and a capsid protein.
Further reports of viruses that appear to have evolved from recombination events between ssRNA and ssDNA viruses have been made.
A new virus has been isolated from the diatom Chaetoceros setoensis. It has a single stranded DNA genome and does not appear to be a member of any previously described group.
A virus - FLIP (Flavobacterium-infecting, lipid-containing phage) - has been isolated from a lake.
This virus has a circular ssDNA genome (9,174 nucleotides) and an
internal lipid membrane enclosed in an icosahedral capsid. The capsid
organisation is he capsid organization pseudo T = 21 dextro. The
major capsid protein has two β-barrels. The capsid organisation is
similar to bacteriophage PM2 - a double stranded bacterial virus.
Satellite viruses
Satellite viruses
are small viruses with either RNA or DNA as their genomic material that
require another virus to replicate. There are two types of DNA
satellite viruses—the alphasatellites and the betasatellites—both of which are dependent on begomoviruses. At present satellite viruses are not classified into genera or higher taxa.
Alphasatellites are small circular single strand DNA viruses that
require a begomovirus for transmission. Betasatellites are small linear
single stranded DNA viruses that require a begomovirus to replicate.
Phylogenetic relationships
Introduction
Phylogenetic
relationships between these families are difficult to determine. The
genomes differ significantly in size and organisation. Most studies that
have attempted to determine these relationships are based either on
some of the more conserved proteins—DNA polymerase and others—or on
common structural features. In general most of the proposed
relationships are tentative and have not yet been used by the ICTV in
their classification.
ds DNA viruses
Herpesviruses and caudoviruses
While
determining the phylogenetic relations between the various known clades
of viruses is difficult, on a number of grounds the herpesviruses and
caudoviruses appear to be related.
While the three families in the order Herpesvirales are clearly
related on morphological grounds, it has proven difficult to determine
the dates of divergence between them because of the lack of gene
conservation. On morphological grounds they appear to be related to the bacteriophages—specifically the Caudoviruses.
The branching order among the herpes viruses suggests that Alloherpesviridae is the basal clade and that Herpesviridae and Malacoherpesviridae are sister clades.
Given the phylogenetic distances between vertebrates and molluscs this
suggests that herpesviruses were initially fish viruses and that they
have evolved with their hosts to infect other vertebrates.
The vertebrate herpes viruses initially evolved ~400 million years ago and underwent subsequent evolution on the supercontinent Pangaea. The alphaherpesvirinae separated from the branch leading to the betaherpesvirinae and gammaherpesvirinae about 180 million years ago to 220 million years ago. The avian herpes viruses diverged from the branch leading to the mammalian species.
The mammalian species divided into two branches—the Simplexvirus and
Varicellovirus genera. This latter divergence appears to have occurred
around the time of the mammalian radiation.
Several dsDNA bacteriophages and the herpesviruses encode a
powerful ATP driven DNA translocating machine that encapsidates a viral
genome into a preformed capsid shell or prohead. The critical components
of the packaging machine are the packaging enzyme (terminase) which
acts as the motor and the portal protein that forms the unique DNA
entrance vertex of prohead. The terminase complex consists of a
recognition subunit (small terminase) and an endonuclease/translocase
subunit (large terminase) and cuts viral genome concatemers. It forms a
motor complex containing five large terminase subunits. The
terminase-viral DNA complex docks on the portal vertex. The pentameric
motor processively translocates DNA until the head shell is full with
one viral genome. The motor cuts the DNA again and dissociates from the
full head, allowing head-finishing proteins to assemble on the portal,
sealing the portal, and constructing a platform for tail attachment.
Only a single gene encoding the putative ATPase subunit of the terminase (UL15) is conserved among all herpesviruses. To a lesser extent this gene is also found in T4-like bacteriophages suggesting a common ancestor for these two groups of viruses. Another paper has also suggested that herpesviruses originated among the bacteriophages.
A common origin for the herpesviruses and the caudoviruses has
been suggested on the basis of parallels in their capsid assembly
pathways and similarities between their portal complexes, through which
DNA enters the capsid.
These two groups of viruses share a distinctive 12-fold arrangement of
subunits in the portal complex. A second paper has suggested an
evolutionary relationship between these two groups of viruses.
It seems likely that the tailed viruses infecting the archaea are also related to the tailed viruses infecting bacteria.
Large DNA viruses
The nucleocytoplasmic large DNA virus group (Asfarviridae, Iridoviridae, Marseilleviridae, Mimiviridae, Phycodnaviridae and Poxviridae) along with three other families—Adenoviridae, Cortiviridae and Tectiviridae—
and the phage Sulfolobus turreted icosahedral virus and the satellite
virus Sputnik all possess double β-barrel major capsid proteins
suggesting a common origin.
Several studies have suggested that the family Ascoviridae evolved from the Iridoviridae.
A study of the Iridoviruses suggests that the Iridoviridae, Ascoviridae
and Marseilleviridae are related with Ascoviruses most closely related
to Iridoviruses.
The family Polydnaviridae may have evolved from the Ascoviridae. Molecular evidence suggests that the Phycodnaviridae may have evolved from the family Iridoviridae. These four families (Ascoviridae, Iridoviridae, Phycodnaviridae and Polydnaviridae) may form a clade but more work is needed to confirm this.
Some of the relations among the large viruses have been established.
Mimiviruses are distantly related to Phycodnaviridae. Pandoraviruses
share a common ancestor with Coccolithoviruses within the family
Phycodnaviridae. Pithoviruses are related to Iridoviridae and Marseilleviridae.
Based on the genome organisation and DNA replication mechanism it
seems that phylogenetic relationships may exist between the rudiviruses
(Rudiviridae) and the large eukaryal DNA viruses: the African swine fever virus (Asfarviridae), Chlorella viruses (Phycodnaviridae) and poxviruses (Poxviridae).
Based on the analysis of the DNA polymerase the genus Dinodnavirus may be a member of the family Asfarviridae. Further work on this virus will required before a final assignment can be made.
It has been suggested that at least some of the giant viruses may originate from mitochondria.
Other viruses
Based on the analysis of the coat protein, Sulfolobus turreted icosahedral virus may share a common ancestry with the Tectiviridae.
The families Adenoviridae and Tectiviridae appear to be related structurally.
Baculoviruses evolved from the nudiviruses 310 million years ago.
The Hytrosaviridae are related to the baculoviruses and to
a lesser extent the nudiviruses suggesting they may have evolved from
the baculoviruses.
The Nimaviridae may be related to nudiviruses and baculoviruses.
The Nudiviruses seem to be related to the polydnaviruses.
A protein common to the families Bicaudaviridae, Lipotrixviridae and Rudiviridae and the unclassified virus Sulfolobus turreted icosahedral virus is known suggesting a common origin.
Examination of the pol genes that encode the DNA dependent DNA polymerase in various groups of viruses suggests a number of possible evolutionary relationships. All know viral DNA polymerases belong to the DNA pol families A and B. All possess a 3'-5'-exonuclease
domain with three sequence motifs Exo I, Exo II and Exo III. The
families A and B are distinguishable with family A Pol sharing 9
distinct consensus sequences and only two of them are convincingly
homologous to sequence motif B of family B. The putative sequence motifs
A, B, and C of the polymerase domain are located near the C-terminus in
family A Pol and more central in family B Pol.
Phylogenetic analysis of these genes places the adenoviruses (Adenoviridae), bacteriophages (Caudovirales)
and the plant and fungal linear plasmids into a single clade. A second
clade includes the alpha- and delta-like viral Pol from insect
ascovirus (Ascoviridae), mammalian herpesviruses (Herpesviridae), fish lymphocystis disease virus (Iridoviridae) and chlorella virus (Phycoviridae). The pol genes of the African swine fever virus (Asfarviridae), baculoviruses (Baculoviridae), fish herpesvirus (Herpesviridae), T-even bacteriophages (Myoviridae) and poxviruses (Poxviridae)
were not clearly resolved. A second study showed that poxvirus,
baculovirus and the animal herpesviruses form separate and distinct
clades. Their relationship to the Asfarviridae and the Myoviridae was not examined and remains unclear.
The polymerases from the archaea are similar to family B DNA Pols. The T4-like viruses infect both bacteria and archaea and their pol gene resembles that of eukaryotes. The DNA polymerase of mitochondria resembles that of the T odd phages (Myoviridae).
The virophage—Mavirus—may have evolved from a recombination between a transposon of the Polinton (Maverick) family and an unknown virus.
The polyoma and papillomaviruses appear to have evolved from single-stranded DNA viruses and ultimately from plasmids.
ss DNA viruses
The
evolutionary history of this group is currently poorly understood. An
ancient origin for the single stranded circular DNA viruses has been
proposed.
Capsid proteins of most icosahedral ssRNA and ssDNA viruses
display the same structural fold, the eight-stranded beta-barrel, also
known as the jelly-roll fold.
On the other hand, the replication proteins of icosahedral ssDNA
viruses belong to the superfamily of rolling-circle replication
initiation proteins that are commonly found in prokaryotic plasmids.
Based on these observations, it has been proposed that small DNA
viruses have originated via recombination between RNA viruses and
plasmids.
Circoviruses may have evolved from a nanovirus.
Given the similarities between the rep proteins of the alphasatellites and the nanoviruses, it is likely that the alphasatellites evolved from the nanoviruses. Further work in this area is needed to clarify this.
The geminiviruses may have evolved from phytoplasmal plasmids. The Genomoviridae and the Geminividiae appear to be related.
Based on the three-dimensional structure of the Rep proteins the geminiviruses and parvoviruses may be related.
The ancestor of the geminiviruses probably infected dicots.
The parvoviruses have frequently invaded the germ lines of diverse animal species including mammals, fishes, birds, tunicates, arthropods and flatworms. In particular they have been associated with the human genome for ~98 million years.
Members of the family Bidnaviridae
have evolved from insect parvoviruses by replacing the typical
replication-initiation endonuclease with a protein-primed family B DNA
polymerase acquired from large DNA transposons of the Polinton/Maverick
family. Some bidnavirus genes were also horizontally acquired from
reoviruses (dsRNA genomes) and baculoviruses (dsDNA genomes).
Bacteriophage evolution
Since 1959 ~6300 prokaryote viruses have been described morphologically, including ~6200 bacterial and ~100 archaeal viruses.
Archaeal viruses belong to 15 families and infect members of 16
archaeal genera. These are nearly exclusively hyperthermophiles or
extreme halophiles. Tailed archaeal viruses are found only in the Euryarchaeota, whereas most filamentous and pleomorphic archaeal viruses occur in the Crenarchaeota. Bacterial viruses belong to 10 families and infect members of 179 bacterial genera: most these are members of the Firmicutes and γ-proteobacteria.
The vast majority (96.3%) are tailed with and only 230 (3.7%) are polyhedral, filamentous or pleomorphic. The family Siphoviridae
is the largest family (>3600 descriptions: 57.3%). The tailed phages
appear to be monophyletic and are the oldest known virus group. They arose repeatedly in different hosts and there are at least 11 separate lines of descent.
All of the known temperate phages employ one of only three
different systems for their lysogenic cycle: lambda-like
integration/excision, Mu-like transposition or the plasmid-like
partitioning of phage N15.
A putative course of evolution of these phages has been proposed by Ackermann.
Tailed phages originated in the early Precambrian, long before eukaryotes and their viruses. The ancestral tailed phage had an icosahedral head of about 60 nanometers in diameter and a long non contractile tail with sixfold symmetry. The capsid
contained a single molecule of double stranded DNA of about 50
kilobases. The tail was probably provided with a fixation apparatus. The
head and tail were held together by a connector. The viral particle
contained no lipids,
was heavier than its descendant viruses and had a high DNA content
proportional to its capsid size (~50%). Most of the genome coded for
structural proteins. Morphopoietic genes clustered at one end of the
genome, with head genes preceding tail genes. Lytic enzymes were
probably coded for. Part of the phage genome was nonessential and
possibly bacterial.
The virus infected its host from the outside and injected its
DNA. Replication involved transcription in several waves and formation
of DNA concatemers.
New phages were released by burst of the infected cell after lysis of host membranes by a peptidoglycan hydrolase.
Capsids were assembled from a starting point, the connector and around a
scaffold. They underwent an elaborate maturation process involving
protein cleavage and capsid expansion. Heads and tails were assembled
separately and joined later. The DNA was cut to size and entered
preformed capsids by a headful mechanism.
Subsequently, the phages evolved contractile or short tails and elongated heads. Some viruses become temperate by acquiring an integrase-excisionase complex, plasmid parts or transposons.
A possible evolutionary pathway using vesicles rather than a protein coat has been described in the archaeal plasmid pR1SE.
NCLDVs
The asfarviruses, iridoviruses, mimiviruses, phycodnaviruses and poxviruses have been shown to belong to a single group,—the large nuclear and cytoplasmic DNA viruses. These are also abbreviated "NCLDV". This clade can be divided into two groups:
- the iridoviruses-phycodnaviruses-mimiviruses group. The phycodnaviruses and mimiviruses are sister clades.
- the poxvirus-asfarviruses group.
It is probable that these viruses evolved before the separation of
eukaryoyes into the extant crown groups. The ancestral genome was
complex with at least 41 genes including (1) the replication machinery
(2) up to four RNA polymerase subunits (3) at least three transcription factors
(4) capping and polyadenylation enzymes (5) the DNA packaging apparatus
(6) and structural components of an icosahedral capsid and the viral
membrane.
The evolution of this group of viruses appears to be complex with genes having been gained from multiple sources.
It has been proposed that the ancestor of NCLDVs has evolved from
large, virus-like DNA transposons of the Polinton/Maverick family.
From Polinton/Maverick transposons NCLDVs might have inherited the key
components required for virion morphogenesis, including the major and
minor capsid proteins, maturation protease and genome packaging ATPase.
Another group of large viruses—the Pandoraviridae—has been described. Two species—Pandoravirus salinus and Pandoravirus dulcis—have been recognized. These were isolated from Chile and Australia respectively. These viruses are about one micrometer
in diameter making them one of the largest viruses discovered so far.
Their gene complement is larger than any other known virus to date. At
present they appear to be unrelated to any other species of virus.
An even larger genus, Pithovirus, has since been discovered, measuring about 1.5 µm in length. Another virus - Cedratvirus - may be related this group.