Bacteria are a type of biological cell. They constitute a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria have a number of shapes, ranging from spheres to rods and spirals. Bacteria were among the first life forms to appear on Earth, and are present in most of its habitats. Bacteria inhabit soil, water, acidic hot springs, radioactive waste, and the deep portions of Earth's crust. Bacteria also live in symbiotic and parasitic relationships with plants and animals. Most bacteria have not been characterised, and only about half of the bacterial phyla have species that can be grown in the laboratory. The study of bacteria is known as bacteriology, a branch of microbiology.
There are typically 40 million bacterial cells in a gram of soil and a million bacterial cells in a millilitre of fresh water. There are approximately 5×1030 bacteria on Earth, forming a biomass which exceeds that of all plants and animals. Bacteria are vital in many stages of the nutrient cycle by recycling nutrients such as the fixation of nitrogen from the atmosphere. The nutrient cycle includes the decomposition of dead bodies; bacteria are responsible for the putrefaction stage in this process. In the biological communities surrounding hydrothermal vents and cold seeps, extremophile bacteria provide the nutrients needed to sustain life by converting dissolved compounds, such as hydrogen sulphide and methane, to energy. Data reported by researchers in October 2012 and published in March 2013 suggested that bacteria thrive in the Mariana Trench, which, with a depth of up to 11 kilometres, is the deepest known part of the oceans. Other researchers reported related studies that microbes thrive inside rocks up to 580 metres below the sea floor under 2.6 kilometres of ocean off the coast of the northwestern United States. According to one of the researchers, "You can find microbes everywhere—they're extremely adaptable to conditions, and survive wherever they are."
The famous notion that bacterial cells in the human body outnumber human cells by a factor of 10:1 has been debunked. There are approximately 39 trillion bacterial cells in the human microbiota as personified by a "reference" 70 kg male 170 cm tall, whereas there are 30 trillion human cells in the body. This means that although they do have the upper hand in actual numbers, it is only by 30%, and not 900%.
The largest number exist in the gut flora, and a large number on the skin. The vast majority of the bacteria in the body are rendered harmless by the protective effects of the immune system, though many are beneficial, particularly in the gut flora. However several species of bacteria are pathogenic and cause infectious diseases, including cholera, syphilis, anthrax, leprosy, and bubonic plague. The most common fatal bacterial diseases are respiratory infections, with tuberculosis alone killing about 2 million people per year, mostly in sub-Saharan Africa. In developed countries, antibiotics are used to treat bacterial infections and are also used in farming, making antibiotic resistance a growing problem. In industry, bacteria are important in sewage treatment and the breakdown of oil spills, the production of cheese and yogurt through fermentation, the recovery of gold, palladium, copper and other metals in the mining sector, as well as in biotechnology, and the manufacture of antibiotics and other chemicals.
Once regarded as plants constituting the class Schizomycetes, bacteria are now classified as prokaryotes. Unlike cells of animals and other eukaryotes, bacterial cells do not contain a nucleus and rarely harbour membrane-bound organelles. Although the term bacteria traditionally included all prokaryotes, the scientific classification changed after the discovery in the 1990s that prokaryotes consist of two very different groups of organisms that evolved from an ancient common ancestor. These evolutionary domains are called Bacteria and Archaea.
Etymology
-4500 —
–
-4000 —
–
-3500 —
–
-3000 —
–
-2500 —
–
-2000 —
–
-1500 —
–
-1000 —
–
-500 —
–
0 —
Origin and early evolution
The ancestors of modern bacteria were unicellular microorganisms that were the first forms of life to appear on Earth, about 4 billion years ago. For about 3 billion years, most organisms were microscopic, and bacteria and archaea were the dominant forms of life. Although bacterial fossils exist, such as stromatolites, their lack of distinctive morphology prevents them from being used to examine the history of bacterial evolution, or to date the time of origin of a particular bacterial species. However, gene sequences can be used to reconstruct the bacterial phylogeny, and these studies indicate that bacteria diverged first from the archaeal/eukaryotic lineage. The most recent common ancestor of bacteria and archaea was probably a hyperthermophile that lived about 2.5 billion–3.2 billion years ago.Bacteria were also involved in the second great evolutionary divergence, that of the archaea and eukaryotes. Here, eukaryotes resulted from the entering of ancient bacteria into endosymbiotic associations with the ancestors of eukaryotic cells, which were themselves possibly related to the Archaea. This involved the engulfment by proto-eukaryotic cells of alphaproteobacterial symbionts to form either mitochondria or hydrogenosomes, which are still found in all known Eukarya (sometimes in highly reduced form, e.g. in ancient "amitochondrial" protozoa). Later, some eukaryotes that already contained mitochondria also engulfed cyanobacteria-like organisms, leading to the formation of chloroplasts in algae and plants. This is known as primary endosymbiosis.
Morphology
Bacteria display many cell morphologies and arrangements
Bacteria display a wide diversity of shapes and sizes, called morphologies. Bacterial cells are about one-tenth the size of eukaryotic cells and are typically 0.5–5.0 micrometres in length. However, a few species are visible to the unaided eye—for example, Thiomargarita namibiensis is up to half a millimetre long and Epulopiscium fishelsoni reaches 0.7 mm. Among the smallest bacteria are members of the genus Mycoplasma, which measure only 0.3 micrometres, as small as the largest viruses. Some bacteria may be even smaller, but these ultramicrobacteria are not well-studied.
Most bacterial species are either spherical, called cocci (sing. coccus, from Greek kókkos, grain, seed), or rod-shaped, called bacilli (sing. bacillus, from Latin baculus, stick). Some bacteria, called vibrio, are shaped like slightly curved rods or comma-shaped; others can be spiral-shaped, called spirilla, or tightly coiled, called spirochaetes. A small number of other unusual shapes have been described, such as star-shaped bacteria. This wide variety of shapes is determined by the bacterial cell wall and cytoskeleton,
and is important because it can influence the ability of bacteria to
acquire nutrients, attach to surfaces, swim through liquids and escape predators.
The range of sizes shown by prokaryotes, relative to those of other organisms and biomolecules.
Many bacterial species exist simply as single cells, others associate in characteristic patterns: Neisseria form diploids (pairs), Streptococcus form chains, and Staphylococcus
group together in "bunch of grapes" clusters. Bacteria can also group
to form larger multicellular structures, such as the elongated filaments of Actinobacteria, the aggregates of Myxobacteria, and the complex hyphae of Streptomyces. These multicellular structures are often only seen in certain conditions. For example, when starved of amino acids, Myxobacteria detect surrounding cells in a process known as quorum sensing,
migrate towards each other, and aggregate to form fruiting bodies up to
500 micrometres long and containing approximately 100,000 bacterial
cells.
In these fruiting bodies, the bacteria perform separate tasks; for
example, about one in ten cells migrate to the top of a fruiting body
and differentiate into a specialised dormant state called a myxospore,
which is more resistant to drying and other adverse environmental
conditions.
Bacteria often attach to surfaces and form dense aggregations called biofilms, and larger formations known as microbial mats.
These biofilms and mats can range from a few micrometres in thickness
to up to half a metre in depth, and may contain multiple species of
bacteria, protists and archaea.
Bacteria living in biofilms display a complex arrangement of cells and
extracellular components, forming secondary structures, such as microcolonies, through which there are networks of channels to enable better diffusion of nutrients. In natural environments, such as soil or the surfaces of plants, the majority of bacteria are bound to surfaces in biofilms.
Biofilms are also important in medicine, as these structures are often
present during chronic bacterial infections or in infections of implanted medical devices, and bacteria protected within biofilms are much harder to kill than individual isolated bacteria.
Cellular structure
Structure and contents of a typical gram-positive bacterial cell (seen by the fact that only one cell membrane is present).
Intracellular structures
The bacterial cell is surrounded by a cell membrane which is made primarily of phospholipids. This membrane encloses the contents of the cell and acts as a barrier to hold nutrients, proteins and other essential components of the cytoplasm within the cell. Unlike eukaryotic cells, bacteria usually lack large membrane-bound structures in their cytoplasm such as a nucleus, mitochondria, chloroplasts and the other organelles present in eukaryotic cells. However, some bacteria have protein-bound organelles in the cytoplasm which compartmentalize aspects of bacterial metabolism, such as the carboxysome. Additionally, bacteria have a multi-component cytoskeleton to control the localisation of proteins and nucleic acids within the cell, and to manage the process of cell division.
Many important biochemical reactions, such as energy generation, occur due to concentration gradients across membranes, creating a potential difference analogous to a battery. The general lack of internal membranes in bacteria means these reactions, such as electron transport, occur across the cell membrane between the cytoplasm and the outside of the cell or periplasm.
However, in many photosynthetic bacteria the plasma membrane is highly
folded and fills most of the cell with layers of light-gathering
membrane. These light-gathering complexes may even form lipid-enclosed structures called chlorosomes in green sulfur bacteria.
An electron micrograph of Halothiobacillus neapolitanus cells with carboxysomes inside, with arrows highlighting visible carboxysomes. Scale bars indicate 100 nm.
Bacteria do not have a membrane-bound nucleus, and their genetic material is typically a single circular bacterial chromosome of DNA located in the cytoplasm in an irregularly shaped body called the nucleoid. The nucleoid contains the chromosome with its associated proteins and RNA. Like all other organisms, bacteria contain ribosomes for the production of proteins, but the structure of the bacterial ribosome is different from that of eukaryotes and Archaea.
Some bacteria produce intracellular nutrient storage granules, such as glycogen, polyphosphate, sulfur or polyhydroxyalkanoates. Certain bacterial species, such as the photosynthetic Cyanobacteria, produce internal gas vacuoles
which they use to regulate their buoyancy, allowing them to move up or
down into water layers with different light intensities and nutrient
levels.
Extracellular structures
Around the outside of the cell membrane is the cell wall. Bacterial cell walls are made of peptidoglycan (also called murein), which is made from polysaccharide chains cross-linked by peptides containing D-amino acids. Bacterial cell walls are different from the cell walls of plants and fungi, which are made of cellulose and chitin, respectively.
The cell wall of bacteria is also distinct from that of Archaea, which
do not contain peptidoglycan. The cell wall is essential to the survival
of many bacteria, and the antibiotic penicillin is able to kill bacteria by inhibiting a step in the synthesis of peptidoglycan.
There are broadly speaking two different types of cell wall in bacteria, that classify bacteria into gram-positive bacteria and gram-negative bacteria. The names originate from the reaction of cells to the Gram stain, a long-standing test for the classification of bacterial species.
Gram-positive bacteria possess a thick cell wall containing many layers of peptidoglycan and teichoic acids.
In contrast, gram-negative bacteria have a relatively thin cell wall
consisting of a few layers of peptidoglycan surrounded by a second lipid membrane containing lipopolysaccharides and lipoproteins. Most bacteria have the gram-negative cell wall, and only the Firmicutes and Actinobacteria
(previously known as the low G+C and high G+C gram-positive bacteria,
respectively) have the alternative gram-positive arrangement. These differences in structure can produce differences in antibiotic susceptibility; for instance, vancomycin can kill only gram-positive bacteria and is ineffective against gram-negative pathogens, such as Haemophilus influenzae or Pseudomonas aeruginosa.
Some bacteria have cell wall structures that are neither classically
gram-positive or gram-negative. This includes clinically important
bacteria such as Mycobacteria which have a thick peptidoglycan cell wall like a gram-positive bacterium, but also a second outer layer of lipids.
In many bacteria, an S-layer of rigidly arrayed protein molecules covers the outside of the cell. This layer provides chemical and physical protection for the cell surface and can act as a macromolecular diffusion barrier. S-layers have diverse but mostly poorly understood functions, but are known to act as virulence factors in Campylobacter and contain surface enzymes in Bacillus stearothermophilus.
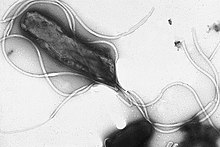
Helicobacter pylori electron micrograph, showing multiple flagella on the cell surface
Flagella are rigid protein structures, about 20 nanometres in diameter and up to 20 micrometres in length, that are used for motility. Flagella are driven by the energy released by the transfer of ions down an electrochemical gradient across the cell membrane.
Fimbriae (sometimes called "attachment pili")
are fine filaments of protein, usually 2–10 nanometres in diameter and
up to several micrometres in length. They are distributed over the
surface of the cell, and resemble fine hairs when seen under the electron microscope.
Fimbriae are believed to be involved in attachment to solid surfaces or
to other cells, and are essential for the virulence of some bacterial
pathogens. Pili (sing. pilus) are cellular appendages, slightly larger than fimbriae, that can transfer genetic material between bacterial cells in a process called conjugation where they are called conjugation pili or sex pili (see bacterial genetics, below). They can also generate movement where they are called type IV pili.
Glycocalyx is produced by many bacteria to surround their cells, and varies in structural complexity: ranging from a disorganised slime layer of extracellular polymeric substances to a highly structured capsule. These structures can protect cells from engulfment by eukaryotic cells such as macrophages (part of the human immune system). They can also act as antigens and be involved in cell recognition, as well as aiding attachment to surfaces and the formation of biofilms.
The assembly of these extracellular structures is dependent on bacterial secretion systems.
These transfer proteins from the cytoplasm into the periplasm or into
the environment around the cell. Many types of secretion systems are
known and these structures are often essential for the virulence of pathogens, so are intensively studied.
Endospores
Bacillus anthracis (stained purple) growing in cerebrospinal fluid
Certain genera of gram-positive bacteria, such as Bacillus, Clostridium, Sporohalobacter, Anaerobacter, and Heliobacterium, can form highly resistant, dormant structures called endospores. Endospores develop within the cytoplasm of the cell; generally a single endospore develops in each cell. Each endospore contains a core of DNA and ribosomes surrounded by a cortex layer and protected by a multilayer rigid coat composed of peptidoglycan and a variety of proteins.
Endospores show no detectable metabolism and can survive extreme physical and chemical stresses, such as high levels of UV light, gamma radiation, detergents, disinfectants, heat, freezing, pressure, and desiccation. In this dormant state, these organisms may remain viable for millions of years, and endospores even allow bacteria to survive exposure to the vacuum and radiation in space. Endospore-forming bacteria can also cause disease: for example, anthrax can be contracted by the inhalation of Bacillus anthracis endospores, and contamination of deep puncture wounds with Clostridium tetani endospores causes tetanus.
Metabolism
Bacteria exhibit an extremely wide variety of metabolic types. The distribution of metabolic traits within a group of bacteria has traditionally been used to define their taxonomy, but these traits often do not correspond with modern genetic classifications. Bacterial metabolism is classified into nutritional groups on the basis of three major criteria: the source of energy, the electron donors used, and the source of carbon used for growth.
Bacteria either derive energy from light using photosynthesis (called phototrophy), or by breaking down chemical compounds using oxidation (called chemotrophy). Chemotrophs use chemical compounds as a source of energy by transferring electrons from a given electron donor to a terminal electron acceptor in a redox reaction.
This reaction releases energy that can be used to drive metabolism.
Chemotrophs are further divided by the types of compounds they use to
transfer electrons. Bacteria that use inorganic compounds such as
hydrogren, carbon monoxide, or ammonia as sources of electrons are called lithotrophs, while those that use organic compounds are called organotrophs. The compounds used to receive electrons are also used to classify bacteria: aerobic organisms use oxygen as the terminal electron acceptor, while anaerobic organisms use other compounds such as nitrate, sulfate, or carbon dioxide.
Many bacteria get their carbon from other organic carbon, called heterotrophy. Others such as cyanobacteria and some purple bacteria are autotrophic, meaning that they obtain cellular carbon by fixing carbon dioxide. In unusual circumstances, the gas methane can be used by methanotrophic bacteria as both a source of electrons and a substrate for carbon anabolism.
| Nutritional type | Source of energy | Source of carbon | Examples |
|---|---|---|---|
| Phototrophs | Sunlight | Organic compounds (photoheterotrophs) or carbon fixation (photoautotrophs) | Cyanobacteria, Green sulfur bacteria, Chloroflexi, or Purple bacteria |
| Lithotrophs | Inorganic compounds | Organic compounds (lithoheterotrophs) or carbon fixation (lithoautotrophs) | Thermodesulfobacteria, Hydrogenophilaceae, or Nitrospirae |
| Organotrophs | Organic compounds | Organic compounds (chemoheterotrophs) or carbon fixation (chemoautotrophs) | Bacillus, Clostridium or Enterobacteriaceae |
In many ways, bacterial metabolism provides traits that are useful
for ecological stability and for human society. One example is that some
bacteria have the ability to fix nitrogen gas using the enzyme nitrogenase. This environmentally important trait can be found in bacteria of most metabolic types listed above. This leads to the ecologically important processes of denitrification, sulfate reduction, and acetogenesis, respectively. Bacterial metabolic processes are also important in biological responses to pollution; for example, sulfate-reducing bacteria are largely responsible for the production of the highly toxic forms of mercury (methyl- and dimethylmercury) in the environment. Non-respiratory anaerobes use fermentation to generate energy and reducing power, secreting metabolic by-products (such as ethanol in brewing) as waste. Facultative anaerobes can switch between fermentation and different terminal electron acceptors depending on the environmental conditions in which they find themselves.
Growth and reproduction
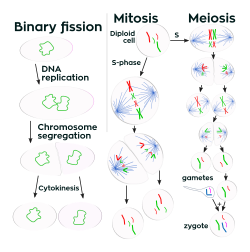
Many bacteria reproduce through binary fission, which is compared to mitosis and meiosis in this image.
Unlike in multicellular organisms, increases in cell size (cell growth) and reproduction by cell division are tightly linked in unicellular organisms. Bacteria grow to a fixed size and then reproduce through binary fission, a form of asexual reproduction.
Under optimal conditions, bacteria can grow and divide extremely
rapidly, and bacterial populations can double as quickly as every
9.8 minutes. In cell division, two identical clone
daughter cells are produced. Some bacteria, while still reproducing
asexually, form more complex reproductive structures that help disperse
the newly formed daughter cells. Examples include fruiting body
formation by Myxobacteria and aerial hyphae formation by Streptomyces, or budding. Budding involves a cell forming a protrusion that breaks away and produces a daughter cell.
A colony of Escherichia coli
In the laboratory, bacteria are usually grown using solid or liquid media. Solid growth media, such as agar plates, are used to isolate
pure cultures of a bacterial strain. However, liquid growth media are
used when measurement of growth or large volumes of cells are required.
Growth in stirred liquid media occurs as an even cell suspension, making
the cultures easy to divide and transfer, although isolating single
bacteria from liquid media is difficult. The use of selective media
(media with specific nutrients added or deficient, or with antibiotics
added) can help identify specific organisms.
Most laboratory techniques for growing bacteria use high levels
of nutrients to produce large amounts of cells cheaply and quickly.
However, in natural environments, nutrients are limited, meaning that
bacteria cannot continue to reproduce indefinitely. This nutrient
limitation has led the evolution of different growth strategies. Some organisms can grow extremely rapidly when nutrients become available, such as the formation of algal (and cyanobacterial) blooms that often occur in lakes during the summer. Other organisms have adaptations to harsh environments, such as the production of multiple antibiotics by Streptomyces that inhibit the growth of competing microorganisms. In nature, many organisms live in communities (e.g., biofilms) that may allow for increased supply of nutrients and protection from environmental stresses. These relationships can be essential for growth of a particular organism or group of organisms (syntrophy).
Bacterial growth
follows four phases. When a population of bacteria first enter a
high-nutrient environment that allows growth, the cells need to adapt to
their new environment. The first phase of growth is the lag phase,
a period of slow growth when the cells are adapting to the
high-nutrient environment and preparing for fast growth. The lag phase
has high biosynthesis rates, as proteins necessary for rapid growth are
produced. The second phase of growth is the logarithmic phase, also known as the exponential phase. The log phase is marked by rapid exponential growth. The rate at which cells grow during this phase is known as the growth rate (k), and the time it takes the cells to double is known as the generation time (g).
During log phase, nutrients are metabolised at maximum speed until one
of the nutrients is depleted and starts limiting growth. The third phase
of growth is the stationary phase
and is caused by depleted nutrients. The cells reduce their metabolic
activity and consume non-essential cellular proteins. The stationary
phase is a transition from rapid growth to a stress response state and
there is increased expression of genes involved in DNA repair, antioxidant metabolism and nutrient transport. The final phase is the death phase where the bacteria run out of nutrients and die.
Genetics
Most bacteria have a single circular chromosome that can range in size from only 160,000 base pairs in the endosymbiotic bacteria Carsonella ruddii, to 12,200,000 base pairs (12.2 Mbp) in the soil-dwelling bacteria Sorangium cellulosum. There are many exceptions to this, for example some Streptomyces and Borrelia species contain a single linear chromosome, while some Vibrio species contain more than one chromosome. Bacteria can also contain plasmids, small extra-chromosomal DNAs that may contain genes for various useful functions such as antibiotic resistance, metabolic capabilities, or various virulence factors.
Bacteria genomes usually encode a few hundred to a few thousand
genes. The genes in bacterial genomes are usually a single continuous
stretch of DNA and although several different types of introns do exist in bacteria, these are much rarer than in eukaryotes.
Bacteria, as asexual organisms, inherit an identical copy of the parent's genomes and are clonal. However, all bacteria can evolve by selection on changes to their genetic material DNA caused by genetic recombination or mutations. Mutations come from errors made during the replication of DNA or from exposure to mutagens.
Mutation rates vary widely among different species of bacteria and even
among different clones of a single species of bacteria.
Genetic changes in bacterial genomes come from either random mutation
during replication or "stress-directed mutation", where genes involved
in a particular growth-limiting process have an increased mutation rate.
Some bacteria also transfer genetic material between cells. This
can occur in three main ways. First, bacteria can take up exogenous DNA
from their environment, in a process called transformation. Many bacteria can naturally take up DNA from the environment, while others must be chemically altered in order to induce them to take up DNA.
The development of competence in nature is usually associated with
stressful environmental conditions, and seems to be an adaptation for
facilitating repair of DNA damage in recipient cells. The second way bacteria transfer genetic material is by transduction, when the integration of a bacteriophage introduces foreign DNA into the chromosome. Many types of bacteriophage exist, some simply infect and lyse their host bacteria, while others insert into the bacterial chromosome. Bacteria resist phage infection through restriction modification systems that degrade foreign DNA, and a system that uses CRISPR
sequences to retain fragments of the genomes of phage that the bacteria
have come into contact with in the past, which allows them to block
virus replication through a form of RNA interference. The third method of gene transfer is conjugation,
whereby DNA is transferred through direct cell contact. In ordinary
circumstances, transduction, conjugation, and transformation involve
transfer of DNA between individual bacteria of the same species, but
occasionally transfer may occur between individuals of different
bacterial species and this may have significant consequences, such as
the transfer of antibiotic resistance. In such cases, gene acquisition from other bacteria or the environment is called horizontal gene transfer and may be common under natural conditions.
Behaviour
Movement
Transmission electron micrograph of Desulfovibrio vulgaris showing a single flagellum at one end of the cell. Scale bar is 0.5 micrometers long.
Many bacteria are motile and can move using a variety of mechanisms. The best studied of these are flagella, long filaments that are turned by a motor at the base to generate propeller-like movement.
The bacterial flagellum is made of about 20 proteins, with
approximately another 30 proteins required for its regulation and
assembly. The flagellum is a rotating structure driven by a reversible motor at the base that uses the electrochemical gradient across the membrane for power.
The different arrangements of bacterial flagella: A-Monotrichous; B-Lophotrichous; C-Amphitrichous; D-Peritrichous
Bacteria can use flagella in different ways to generate different kinds of movement. Many bacteria (such as E. coli)
have two distinct modes of movement: forward movement (swimming) and
tumbling. The tumbling allows them to reorient and makes their movement a
three-dimensional random walk. Bacterial species differ in the number and arrangement of flagella on their surface; some have a single flagellum (monotrichous), a flagellum at each end (amphitrichous), clusters of flagella at the poles of the cell (lophotrichous), while others have flagella distributed over the entire surface of the cell (peritrichous). The flagella of a unique group of bacteria, the spirochaetes, are found between two membranes in the periplasmic space. They have a distinctive helical body that twists about as it moves.
Two other types of bacterial motion are called twitching motility that relies on a structure called the type IV pilus, and gliding motility,
that uses other mechanisms. In twitching motility, the rod-like pilus
extends out from the cell, binds some substrate, and then retracts,
pulling the cell forward.
Motile bacteria are attracted or repelled by certain stimuli in behaviours called taxes: these include chemotaxis, phototaxis, energy taxis, and magnetotaxis. In one peculiar group, the myxobacteria, individual bacteria move together to form waves of cells that then differentiate to form fruiting bodies containing spores. The myxobacteria move only when on solid surfaces, unlike E. coli, which is motile in liquid or solid media.
Several Listeria and Shigella species move inside host cells by usurping the cytoskeleton, which is normally used to move organelles inside the cell. By promoting actin polymerisation at one pole of their cells, they can form a kind of tail that pushes them through the host cell's cytoplasm.
Communication
A few bacteria have chemical systems that generate light. This bioluminescence
often occurs in bacteria that live in association with fish, and the
light probably serves to attract fish or other large animals.
Bacteria often function as multicellular aggregates known as biofilms, exchanging a variety of molecular signals for inter-cell communication, and engaging in coordinated multicellular behaviour.
The communal benefits of multicellular cooperation include a
cellular division of labour, accessing resources that cannot effectively
be used by single cells, collectively defending against antagonists,
and optimising population survival by differentiating into distinct cell
types. For example, bacteria in biofilms can have more than 500 times increased resistance to antibacterial agents than individual "planktonic" bacteria of the same species.
One type of inter-cellular communication by a molecular signal is called quorum sensing,
which serves the purpose of determining whether there is a local
population density that is sufficiently high that it is productive to
invest in processes that are only successful if large numbers of similar
organisms behave similarly, as in excreting digestive enzymes or
emitting light.
Quorum sensing allows bacteria to coordinate gene expression, and enables them to produce, release and detect autoinducers or pheromones which accumulate with the growth in cell population.
Classification and identification
Streptococcus mutans visualised with a Gram stain
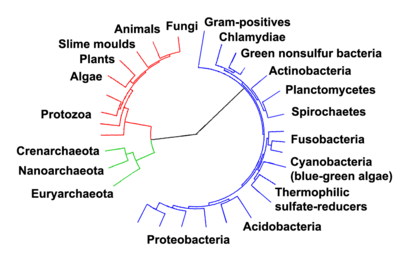
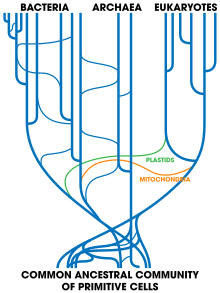
Phylogenetic tree showing the diversity of bacteria, compared to other organisms. Eukaryotes are coloured red, archaea green and bacteria blue.
Classification seeks to describe the diversity of bacterial species by naming and grouping organisms based on similarities. Bacteria can be classified on the basis of cell structure, cellular metabolism or on differences in cell components, such as DNA, fatty acids, pigments, antigens and quinones. While these schemes allowed the identification and classification of bacterial strains, it was unclear whether these differences represented variation between distinct species or between strains of the same species. This uncertainty was due to the lack of distinctive structures in most bacteria, as well as lateral gene transfer between unrelated species. Due to lateral gene transfer, some closely related bacteria can have very different morphologies and metabolisms. To overcome this uncertainty, modern bacterial classification emphasises molecular systematics, using genetic techniques such as guanine cytosine ratio determination, genome-genome hybridisation, as well as sequencing genes that have not undergone extensive lateral gene transfer, such as the rRNA gene. Classification of bacteria is determined by publication in the International Journal of Systematic Bacteriology, and Bergey's Manual of Systematic Bacteriology. The International Committee on Systematic Bacteriology (ICSB) maintains international rules for the naming of bacteria and taxonomic categories and for the ranking of them in the International Code of Nomenclature of Bacteria.
The term "bacteria" was traditionally applied to all microscopic,
single-cell prokaryotes. However, molecular systematics showed
prokaryotic life to consist of two separate domains, originally called Eubacteria and Archaebacteria, but now called Bacteria and Archaea that evolved independently from an ancient common ancestor.
The archaea and eukaryotes are more closely related to each other than
either is to the bacteria. These two domains, along with Eukarya, are
the basis of the three-domain system, which is currently the most widely used classification system in microbiology.
However, due to the relatively recent introduction of molecular
systematics and a rapid increase in the number of genome sequences that
are available, bacterial classification remains a changing and expanding
field. For example, a few biologists argue that the Archaea and Eukaryotes evolved from gram-positive bacteria.
The identification of bacteria in the laboratory is particularly relevant in medicine,
where the correct treatment is determined by the bacterial species
causing an infection. Consequently, the need to identify human pathogens
was a major impetus for the development of techniques to identify
bacteria.
The Gram stain, developed in 1884 by Hans Christian Gram, characterises bacteria based on the structural characteristics of their cell walls.
The thick layers of peptidoglycan in the "gram-positive" cell wall
stain purple, while the thin "gram-negative" cell wall appears pink. By
combining morphology and Gram-staining, most bacteria can be classified
as belonging to one of four groups (gram-positive cocci, gram-positive
bacilli, gram-negative cocci and gram-negative bacilli). Some organisms
are best identified by stains other than the Gram stain, particularly
mycobacteria or Nocardia, which show acid-fastness on Ziehl–Neelsen or similar stains. Other organisms may need to be identified by their growth in special media, or by other techniques, such as serology.
Culture
techniques are designed to promote the growth and identify particular
bacteria, while restricting the growth of the other bacteria in the
sample. Often these techniques are designed for specific specimens; for
example, a sputum sample will be treated to identify organisms that cause pneumonia, while stool specimens are cultured on selective media to identify organisms that cause diarrhoea, while preventing growth of non-pathogenic bacteria. Specimens that are normally sterile, such as blood, urine or spinal fluid, are cultured under conditions designed to grow all possible organisms. Once a pathogenic organism has been isolated, it can be further characterised by its morphology, growth patterns (such as aerobic or anaerobic growth), patterns of hemolysis, and staining.
As with bacterial classification, identification of bacteria is
increasingly using molecular methods. Diagnostics using DNA-based tools,
such as polymerase chain reaction, are increasingly popular due to their specificity and speed, compared to culture-based methods. These methods also allow the detection and identification of "viable but nonculturable" cells that are metabolically active but non-dividing.
However, even using these improved methods, the total number of
bacterial species is not known and cannot even be estimated with any
certainty. Following present classification, there are a little less
than 9,300 known species of prokaryotes, which includes bacteria and
archaea; but attempts to estimate the true number of bacterial diversity have ranged from 107 to 109 total species—and even these diverse estimates may be off by many orders of magnitude.
Interactions with other organisms
Overview of bacterial infections and main species involved.
Despite their apparent simplicity, bacteria can form complex associations with other organisms. These symbiotic associations can be divided into parasitism, mutualism and commensalism.
Due to their small size, commensal bacteria are ubiquitous and grow on
animals and plants exactly as they will grow on any other surface.
However, their growth can be increased by warmth and sweat, and large populations of these organisms in humans are the cause of body odour.
Predators
Some species of bacteria kill and then consume other microorganisms, these species are called predatory bacteria. These include organisms such as Myxococcus xanthus, which forms swarms of cells that kill and digest any bacteria they encounter. Other bacterial predators either attach to their prey in order to digest them and absorb nutrients, such as Vampirovibrio chlorellavorus, or invade another cell and multiply inside the cytosol, such as Daptobacter. These predatory bacteria are thought to have evolved from saprophages that consumed dead microorganisms, through adaptations that allowed them to entrap and kill other organisms.
Mutualists
Certain bacteria form close spatial associations that are essential
for their survival. One such mutualistic association, called
interspecies hydrogen transfer, occurs between clusters of anaerobic bacteria that consume organic acids, such as butyric acid or propionic acid, and produce hydrogen, and methanogenic Archaea that consume hydrogen.
The bacteria in this association are unable to consume the organic
acids as this reaction produces hydrogen that accumulates in their
surroundings. Only the intimate association with the hydrogen-consuming
Archaea keeps the hydrogen concentration low enough to allow the
bacteria to grow.
In soil, microorganisms that reside in the rhizosphere (a zone that includes the root surface and the soil that adheres to the root after gentle shaking) carry out nitrogen fixation, converting nitrogen gas to nitrogenous compounds.
This serves to provide an easily absorbable form of nitrogen for many
plants, which cannot fix nitrogen themselves. Many other bacteria are
found as symbionts in humans and other organisms. For example, the presence of over 1,000 bacterial species in the normal human gut flora of the intestines can contribute to gut immunity, synthesise vitamins, such as folic acid, vitamin K and biotin, convert sugars to lactic acid (see Lactobacillus), as well as fermenting complex undigestible carbohydrates. The presence of this gut flora also inhibits the growth of potentially pathogenic bacteria (usually through competitive exclusion) and these beneficial bacteria are consequently sold as probiotic dietary supplements.
Pathogens
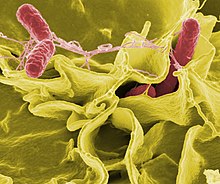
Colour-enhanced scanning electron micrograph showing Salmonella typhimurium (red) invading cultured human cells
If bacteria form a parasitic association with other organisms, they
are classed as pathogens. Pathogenic bacteria are a major cause of human
death and disease and cause infections such as tetanus, typhoid fever, diphtheria, syphilis, cholera, foodborne illness, leprosy and tuberculosis. A pathogenic cause for a known medical disease may only be discovered many years after, as was the case with Helicobacter pylori and peptic ulcer disease. Bacterial diseases are also important in agriculture, with bacteria causing leaf spot, fire blight and wilts in plants, as well as Johne's disease, mastitis, salmonella and anthrax in farm animals.
Each species of pathogen has a characteristic spectrum of interactions with its human hosts. Some organisms, such as Staphylococcus or Streptococcus, can cause skin infections, pneumonia, meningitis and even overwhelming sepsis, a systemic inflammatory response producing shock, massive vasodilation and death. Yet these organisms are also part of the normal human flora and usually exist on the skin or in the nose without causing any disease at all. Other organisms invariably cause disease in humans, such as the Rickettsia, which are obligate intracellular parasites able to grow and reproduce only within the cells of other organisms. One species of Rickettsia causes typhus, while another causes Rocky Mountain spotted fever. Chlamydia, another phylum of obligate intracellular parasites, contains species that can cause pneumonia, or urinary tract infection and may be involved in coronary heart disease. Finally, some species, such as Pseudomonas aeruginosa, Burkholderia cenocepacia, and Mycobacterium avium, are opportunistic pathogens and cause disease mainly in people suffering from immunosuppression or cystic fibrosis.
Bacterial infections may be treated with antibiotics, which are classified as bacteriocidal if they kill bacteria, or bacteriostatic if they just prevent bacterial growth. There are many types of antibiotics and each class inhibits
a process that is different in the pathogen from that found in the
host. An example of how antibiotics produce selective toxicity are chloramphenicol and puromycin, which inhibit the bacterial ribosome, but not the structurally different eukaryotic ribosome. Antibiotics are used both in treating human disease and in intensive farming to promote animal growth, where they may be contributing to the rapid development of antibiotic resistance in bacterial populations. Infections can be prevented by antiseptic
measures such as sterilising the skin prior to piercing it with the
needle of a syringe, and by proper care of indwelling catheters.
Surgical and dental instruments are also sterilised to prevent contamination by bacteria. Disinfectants such as bleach are used to kill bacteria or other pathogens on surfaces to prevent contamination and further reduce the risk of infection.
Significance in technology and industry
Bacteria, often lactic acid bacteria, such as Lactobacillus and Lactococcus, in combination with yeasts and moulds, have been used for thousands of years in the preparation of fermented foods, such as cheese, pickles, soy sauce, sauerkraut, vinegar, wine and yogurt.
The ability of bacteria to degrade a variety of organic compounds is remarkable and has been used in waste processing and bioremediation. Bacteria capable of digesting the hydrocarbons in petroleum are often used to clean up oil spills. Fertiliser was added to some of the beaches in Prince William Sound in an attempt to promote the growth of these naturally occurring bacteria after the 1989 Exxon Valdez oil spill. These efforts were effective on beaches that were not too thickly covered in oil. Bacteria are also used for the bioremediation of industrial toxic wastes. In the chemical industry, bacteria are most important in the production of enantiomerically pure chemicals for use as pharmaceuticals or agrichemicals.
Bacteria can also be used in the place of pesticides in the biological pest control. This commonly involves Bacillus thuringiensis (also called BT), a gram-positive, soil dwelling bacterium. Subspecies of this bacteria are used as a Lepidopteran-specific insecticides under trade names such as Dipel and Thuricide. Because of their specificity, these pesticides are regarded as environmentally friendly, with little or no effect on humans, wildlife, pollinators and most other beneficial insects.
Because of their ability to quickly grow and the relative ease
with which they can be manipulated, bacteria are the workhorses for the
fields of molecular biology, genetics and biochemistry. By making mutations in bacterial DNA and examining the resulting phenotypes, scientists can determine the function of genes, enzymes and metabolic pathways in bacteria, then apply this knowledge to more complex organisms. This aim of understanding the biochemistry of a cell reaches its most complex expression in the synthesis of huge amounts of enzyme kinetic and gene expression data into mathematical models of entire organisms. This is achievable in some well-studied bacteria, with models of Escherichia coli metabolism now being produced and tested. This understanding of bacterial metabolism and genetics allows the use of biotechnology to bioengineer bacteria for the production of therapeutic proteins, such as insulin, growth factors, or antibodies.
Because of their importance for research in general, samples of bacterial strains are isolated and preserved in Biological Resource Centers. This ensures the availability of the strain to scientists worldwide.
History of bacteriology
Antonie van Leeuwenhoek, the first microbiologist and the first person to observe bacteria using a microscope.
Bacteria were first observed by the Dutch microscopist Antonie van Leeuwenhoek in 1676, using a single-lens microscope of his own design. He then published his observations in a series of letters to the Royal Society of London.
Bacteria were Leeuwenhoek's most remarkable microscopic discovery. They
were just at the limit of what his simple lenses could make out and, in
one of the most striking hiatuses in the history of science, no one
else would see them again for over a century. His observations had also included protozoans which he called animalcules, and his findings were looked at again in the light of the more recent findings of cell theory.
Christian Gottfried Ehrenberg introduced the word "bacterium" in 1828. In fact, his Bacterium was a genus that contained non-spore-forming rod-shaped bacteria, as opposed to Bacillus, a genus of spore-forming rod-shaped bacteria defined by Ehrenberg in 1835.
Louis Pasteur demonstrated in 1859 that the growth of microorganisms causes the fermentation process, and that this growth is not due to spontaneous generation. (Yeasts and moulds, commonly associated with fermentation, are not bacteria, but rather fungi.) Along with his contemporary Robert Koch, Pasteur was an early advocate of the germ theory of disease.
Robert Koch, a pioneer in medical microbiology, worked on cholera, anthrax and tuberculosis. In his research into tuberculosis Koch finally proved the germ theory, for which he received a Nobel Prize in 1905. In Koch's postulates, he set out criteria to test if an organism is the cause of a disease, and these postulates are still used today.
Ferdinand Cohn
is said to be a founder of bacteriology, studying bacteria from 1870.
Cohn was the first to classify bacteria based on their morphology.
Though it was known in the nineteenth century that bacteria are the cause of many diseases, no effective antibacterial treatments were available. In 1910, Paul Ehrlich developed the first antibiotic, by changing dyes that selectively stained Treponema pallidum—the spirochaete that causes syphilis—into compounds that selectively killed the pathogen. Ehrlich had been awarded a 1908 Nobel Prize for his work on immunology, and pioneered the use of stains to detect and identify bacteria, with his work being the basis of the Gram stain and the Ziehl–Neelsen stain.
A major step forward in the study of bacteria came in 1977 when Carl Woese recognised that archaea have a separate line of evolutionary descent from bacteria. This new phylogenetic taxonomy depended on the sequencing of 16S ribosomal RNA, and divided prokaryotes into two evolutionary domains, as part of the three-domain system.
Computer-made form
In April 2019, scientists at ETH Zurich reported the creation of the world's first bacterial genome, named Caulobacter ethensis-2.0, made entirely by a computer, although a related viable form of C. ethensis-2.0 does not yet exist.