The RNA world is a hypothetical stage in the evolutionary history of life on Earth, in which self-replicating RNA molecules proliferated before the evolution of DNA and proteins. The term also refers to the hypothesis that posits the existence of this stage.
Alexander Rich first proposed the concept of the RNA world in 1962, and Walter Gilbert coined the term in 1986. Alternative chemical paths to life have been proposed, and RNA-based life may not have been the first life to exist. Even so, the evidence for an RNA world is strong enough that the hypothesis has gained wide acceptance. The concurrent formation of all four RNA building blocks further strengthened the hypothesis.
Like DNA, RNA can store and replicate genetic information; like protein enzymes, RNA enzymes (ribozymes) can catalyze (start or accelerate) chemical reactions that are critical for life. One of the most critical components of cells, the ribosome, is composed primarily of RNA. Ribonucleotide moieties in many coenzymes, such as acetyl-CoA, NADH, FADH, and F420, may be surviving remnants of covalently bound coenzymes in an RNA world.
Although RNA is fragile, some ancient RNAs may have evolved the ability to methylate other RNAs to protect them.
If the RNA world existed, it was probably followed by an age characterized by the evolution of ribonucleoproteins (RNP world), which in turn ushered in the era of DNA and longer proteins. DNA has better stability and durability than RNA; this may explain why it became the predominant information storage molecule. Protein enzymes may have come to replace RNA-based ribozymes as biocatalysts because their greater abundance and diversity of monomers makes them more versatile. As some co-factors contain both nucleotide and amino-acid characteristics, it may be that amino acids, peptides and finally proteins initially were co-factors for ribozymes.
History
One of the challenges in studying abiogenesis is that the system of reproduction and metabolism utilized by all extant life involves three distinct types of interdependent macromolecules (DNA, RNA, and protein). This suggests that life could not have arisen in its current form, which has led researchers to hypothesize mechanisms whereby the current system might have arisen from a simpler precursor system. The concept of RNA as a primordial molecule can be found in papers by Francis Crick and Leslie Orgel, as well as in Carl Woese's 1967 book The Genetic Code. In 1962, the molecular biologist Alexander Rich posited much the same idea in an article he contributed to a volume issued in honor of Nobel-laureate physiologist Albert Szent-Györgyi. Hans Kuhn in 1972 laid out a possible process by which the modern genetic system might have arisen from a nucleotide-based precursor, and this led Harold White in 1976 to observe that many of the cofactors essential for enzymatic function are either nucleotides or could have been derived from nucleotides. He proposed a scenario whereby the critical electrochemistry of enzymatic reactions would have necessitated retention of the specific nucleotide moieties of the original RNA-based enzymes carrying out the reactions, while the remaining structural elements of the enzymes were gradually replaced by protein, until all that remained of the original RNAs were these nucleotide cofactors, "fossils of nucleic acid enzymes". The phrase "RNA World" was first used by Nobel laureate Walter Gilbert in 1986, in a commentary on how recent observations of the catalytic properties of various forms of RNA fit with this hypothesis.
Properties of RNA
The properties of RNA make the idea of the RNA world hypothesis conceptually plausible, though its general acceptance as an explanation for the origin of life requires further evidence. RNA is known to form efficient catalysts and its similarity to DNA makes clear its ability to store information. Opinions differ, however, as to whether RNA constituted the first autonomous self-replicating system or was a derivative of a still-earlier system. One version of the hypothesis is that a different type of nucleic acid, termed pre-RNA, was the first one to emerge as a self-reproducing molecule, to be replaced by RNA only later. On the other hand, the discovery in 2009 that activated pyrimidine ribonucleotides can be synthesized under plausible prebiotic conditions suggests that it is premature to dismiss the RNA-first scenarios. Suggestions for 'simple' pre-RNA nucleic acids have included peptide nucleic acid (PNA), threose nucleic acid (TNA) or glycol nucleic acid (GNA). Despite their structural simplicity and possession of properties comparable with RNA, the chemically plausible generation of "simpler" nucleic acids under prebiotic conditions has yet to be demonstrated.
RNA as an enzyme
RNA enzymes, or ribozymes, are found in today's DNA-based life and could be examples of living fossils. Ribozymes play vital roles, such as that of the ribosome. The large subunit of 70s Ribosome (50s) contains 23s rRNA which act as a peptide bond forming enzyme called peptidal transferase and helps in protein synthesis. Many other ribozyme functions exist; for example, the hammerhead ribozyme performs self-cleavage and an RNA polymerase ribozyme can synthesize a short RNA strand from a primed RNA template.
Among the enzymatic properties important for the beginning of life are:
- Self-replication
- The ability to self-replicate, or synthesize other RNA molecules; relatively short RNA molecules that can synthesize others have been artificially produced in the lab. The shortest was 165 bases long, though it has been estimated that only part of the molecule was crucial for this function. One version, 189 bases long, had an error rate of just 1.1% per nucleotide when synthesizing an 11 nucleotide long RNA strand from primed template strands. This 189 base pair ribozyme could polymerize a template of at most 14 nucleotides in length, which is too short for self replication, but is a potential lead for further investigation. The longest primer extension performed by a ribozyme polymerase was 20 bases. In 2016, researchers reported the use of in vitro evolution to improve dramatically the activity and generality of an RNA polymerase ribozyme by selecting variants that can synthesize functional RNA molecules from an RNA template. Each RNA polymerase ribozyme was engineered to remain linked to its new, synthesized RNA strand; this allowed the team to isolate successful polymerases. The isolated RNA polymerases were again used for another round of evolution. After several rounds of evolution, they obtained one RNA polymerase ribozyme called 24-3 that was able to copy almost any other RNA, from small catalysts to long RNA-based enzymes. Particular RNAs were amplified up to 10,000 times, a first RNA version of the polymerase chain reaction (PCR).
- Catalysis
- The ability to catalyze simple chemical reactions—which would enhance creation of molecules that are building blocks of RNA molecules (i.e., a strand of RNA that would make creating more strands of RNA easier). Relatively short RNA molecules with such abilities have been artificially formed in the lab. A recent study showed that almost any nucleic acid can evolve into a catalytic sequence under appropriate selection. For instance, an arbitrarily chosen 50-nucleotide DNA fragment encoding for the Bos taurus (cattle) albumin mRNA was subjected to test-tube evolution to derive a catalytic DNA (Deoxyribozyme, also called DNAzyme) with RNA-cleavage activity. After only a few weeks, a DNAzyme with significant catalytic activity had evolved. In general, DNA is much more chemically inert than RNA and hence much more resistant to obtaining catalytic properties. If in vitro evolution works for DNA it will happen much more easily with RNA.
- Amino acid-RNA ligation
- The ability to conjugate an amino acid to the 3'-end of an RNA in order to use its chemical groups or provide a long-branched aliphatic side-chain.
- Peptide bond formation
- The ability to catalyse the formation of peptide bonds between amino acids to produce short peptides or longer proteins. This is done in modern cells by ribosomes, a complex of several RNA molecules known as rRNA together with many proteins. The rRNA molecules are thought responsible for its enzymatic activity, as no amino acid molecules lie within 18Å of the enzyme's active site, and, when the majority of the amino acids in the ribosome were stringently removed, the resulting ribosome retained its full peptidyl transferase activity, fully able to catalyze the formation of peptide bonds between amino acids. A much shorter RNA molecule has been synthesized in the laboratory with the ability to form peptide bonds, and it has been suggested that rRNA has evolved from a similar molecule. It has also been suggested that amino acids may have initially been involved with RNA molecules as cofactors enhancing or diversifying their enzymatic capabilities, before evolving into more complex peptides. Similarly, tRNA is suggested to have evolved from RNA molecules that began to catalyze amino acid transfer.
RNA in information storage
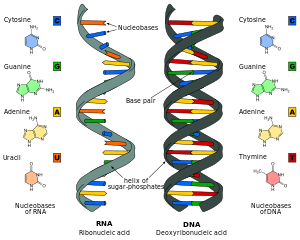
RNA is a very similar molecule to DNA, with only two major chemical differences (the backbone of RNA uses ribose instead of deoxyribose and its nucleobases include uracil instead of thymine). The overall structure of RNA and DNA are immensely similar—one strand of DNA and one of RNA can bind to form a double helical structure. This makes the storage of information in RNA possible in a very similar way to the storage of information in DNA. However, RNA is less stable, being more prone to hydrolysis due to the presence of a hydroxyl group at the ribose 2' position.
Comparison of DNA and RNA structure
The major difference between RNA and DNA is the presence of a hydroxyl group at the 2'-position of the ribose sugar in RNA (illustration, right). This group makes the molecule less stable because, when not constrained in a double helix, the 2' hydroxyl can chemically attack the adjacent phosphodiester bond to cleave the phosphodiester backbone. The hydroxyl group also forces the ribose into the C3'-endo sugar conformation unlike the C2'-endo conformation of the deoxyribose sugar in DNA. This forces an RNA double helix to change from a B-DNA structure to one more closely resembling A-DNA.
RNA also uses a different set of bases than DNA—adenine, guanine, cytosine and uracil, instead of adenine, guanine, cytosine and thymine. Chemically, uracil is similar to thymine, differing only by a methyl group, and its production requires less energy. In terms of base pairing, this has no effect. Adenine readily binds uracil or thymine. Uracil is, however, one product of damage to cytosine that makes RNA particularly susceptible to mutations that can replace a GC base pair with a GU (wobble) or AU base pair.
RNA is thought to have preceded DNA, because of their ordering in the biosynthetic pathways. The deoxyribonucleotides used to make DNA are made from ribonucleotides, the building blocks of RNA, by removing the 2'-hydroxyl group. As a consequence a cell must have the ability to make RNA before it can make DNA.
Limitations of information storage in RNA
The chemical properties of RNA make large RNA molecules inherently fragile, and they can easily be broken down into their constituent nucleotides through hydrolysis. These limitations do not make use of RNA as an information storage system impossible, simply energy intensive (to repair or replace damaged RNA molecules) and prone to mutation. While this makes it unsuitable for current 'DNA optimised' life, it may have been acceptable for more primitive life.
RNA as a regulator
Riboswitches have been found to act as regulators of gene expression, particularly in bacteria, but also in plants and archaea. Riboswitches alter their secondary structure in response to the binding of a metabolite. This change in structure can result in the formation or disruption of a terminator, truncating or permitting transcription respectively. Alternatively, riboswitches may bind or occlude the Shine–Dalgarno sequence, affecting translation. It has been suggested that these originated in an RNA-based world. In addition, RNA thermometers regulate gene expression in response to temperature changes.
Support and difficulties
The RNA world hypothesis is supported by RNA's ability to store, transmit, and duplicate genetic information, as DNA does. RNA can act as a ribozyme, a special type of enzyme. Because it can perform the tasks of both DNA and enzymes, RNA is believed to have once been capable of supporting independent life forms. Some viruses use RNA as their genetic material, rather than DNA. Further, while nucleotides were not found in experiments based on Miller-Urey experiment, their formation in prebiotically plausible conditions was reported in 2009; the purine base known as adenine is merely a pentamer of hydrogen cyanide. Experiments with basic ribozymes, like Bacteriophage Qβ RNA, have shown that simple self-replicating RNA structures can withstand even strong selective pressures (e.g., opposite-chirality chain terminators).
Since there were no known chemical pathways for the abiogenic synthesis of nucleotides from pyrimidine nucleobases cytosine and uracil under prebiotic conditions, it is thought by some that nucleic acids did not contain these nucleobases seen in life's nucleic acids. The nucleoside cytosine has a half-life in isolation of 19 days at 100 °C (212 °F) and 17,000 years in freezing water, which some argue is too short on the geologic time scale for accumulation. Others have questioned whether ribose and other backbone sugars could be stable enough to be found in the original genetic material, and have raised the issue that all ribose molecules would have had to be the same enantiomer, as any nucleotide of the wrong chirality acts as a chain terminator.
Pyrimidine ribonucleosides and their respective nucleotides have been prebiotically synthesised by a sequence of reactions that by-pass free sugars and assemble in a stepwise fashion by including nitrogenous and oxygenous chemistries. In a series of publications, John Sutherland and his team at the School of Chemistry, University of Manchester, have demonstrated high yielding routes to cytidine and uridine ribonucleotides built from small 2- and 3-carbon fragments such as glycolaldehyde, glyceraldehyde or glyceraldehyde-3-phosphate, cyanamide, and cyanoacetylene. One of the steps in this sequence allows the isolation of enantiopure ribose aminooxazoline if the enantiomeric excess of glyceraldehyde is 60% or greater, of possible interest toward biological homochirality. This can be viewed as a prebiotic purification step, where the said compound spontaneously crystallised out from a mixture of the other pentose aminooxazolines. Aminooxazolines can react with cyanoacetylene in a mild and highly efficient manner, controlled by inorganic phosphate, to give the cytidine ribonucleotides. Photoanomerization with UV light allows for inversion about the 1' anomeric centre to give the correct beta stereochemistry; one problem with this chemistry is the selective phosphorylation of alpha-cytidine at the 2' position. However, in 2009, they showed that the same simple building blocks allow access, via phosphate controlled nucleobase elaboration, to 2',3'-cyclic pyrimidine nucleotides directly, which are known to be able to polymerise into RNA. Organic chemist Donna Blackmond described this finding as "strong evidence" in favour of the RNA world. However, John Sutherland said that while his team's work suggests that nucleic acids played an early and central role in the origin of life, it did not necessarily support the RNA world hypothesis in the strict sense, which he described as a "restrictive, hypothetical arrangement".
The Sutherland group's 2009 paper also highlighted the possibility for the photo-sanitization of the pyrimidine-2',3'-cyclic phosphates. A potential weakness of these routes is the generation of enantioenriched glyceraldehyde, or its 3-phosphate derivative (glyceraldehyde prefers to exist as its keto tautomer dihydroxyacetone).
On August 8, 2011, a report, based on NASA studies with meteorites found on Earth, was published suggesting building blocks of RNA (adenine, guanine, and related organic molecules) may have been formed extraterrestrially in outer space. In 2017, a numerical model suggests that the RNA world may have emerged in warm ponds on the early Earth, and that meteorites were a plausible and probable source of the RNA building blocks (ribose and nucleic acids) to these environments. On August 29, 2012, astronomers at Copenhagen University reported the detection of a specific sugar molecule, glycolaldehyde, in a distant star system. The molecule was found around the protostellar binary IRAS 16293-2422, which is located 400 light years from Earth. Because glycolaldehyde is needed to form RNA, this finding suggests that complex organic molecules may form in stellar systems prior to the formation of planets, eventually arriving on young planets early in their formation.
Prebiotic RNA synthesis
Nucleotides are the fundamental molecules that combine in series to form RNA. They consist of a nitrogenous base attached to a sugar-phosphate backbone. RNA is made of long stretches of specific nucleotides arranged so that their sequence of bases carries information. The RNA world hypothesis holds that in the primordial soup (or sandwich), there existed free-floating nucleotides. These nucleotides regularly formed bonds with one another, which often broke because the change in energy was so low. However, certain sequences of base pairs have catalytic properties that lower the energy of their chain being created, enabling them to stay together for longer periods of time. As each chain grew longer, it attracted more matching nucleotides faster, causing chains to now form faster than they were breaking down.
These chains have been proposed by some as the first, primitive forms of life. In an RNA world, different sets of RNA strands would have had different replication outputs, which would have increased or decreased their frequency in the population, i.e. natural selection. As the fittest sets of RNA molecules expanded their numbers, novel catalytic properties added by mutation, which benefitted their persistence and expansion, could accumulate in the population. Such an autocatalytic set of ribozymes, capable of self replication in about an hour, has been identified. It was produced by molecular competition (in vitro evolution) of candidate enzyme mixtures.
Competition between RNA may have favored the emergence of cooperation between different RNA chains, opening the way for the formation of the first protocell. Eventually, RNA chains developed with catalytic properties that help amino acids bind together (a process called peptide-bonding). These amino acids could then assist with RNA synthesis, giving those RNA chains that could serve as ribozymes the selective advantage. The ability to catalyze one step in protein synthesis, aminoacylation of RNA, has been demonstrated in a short (five-nucleotide) segment of RNA.
In March 2015, NASA scientists reported that, for the first time, complex DNA and RNA organic compounds of life, including uracil, cytosine, and thymine, have been formed in the laboratory under conditions found only in outer space, using starting chemicals, like pyrimidine, found in meteorites. Pyrimidine, like polycyclic aromatic hydrocarbons (PAHs), may have been formed in giant red stars or in interstellar dust and gas clouds, according to the scientists.
In 2018, researchers at Georgia Institute of Technology identified three molecular candidates for the bases that might have formed an earliest version of proto-RNA: barbituric acid, melamine, and 2,4,6-triaminopyrimidine (TAP). These three molecules are simpler versions of the four bases in current RNA, which could have been present in larger amounts and could still be forward-compatible with them, but may have been discarded by evolution in exchange for more optimal base pairs. Specifically, TAP can form nucleotides with a large range of sugars. Both TAP and melamine base pair with barbituric acid. All three spontaneously form nucleotides with ribose.
Evolution of DNA
One of the challenges posed by the RNA world hypothesis is to discover the pathway by which an RNA-based system transitioned to one based on DNA. Geoffrey Diemer and Ken Stedman, at Portland State University in Oregon, may have found a solution. While conducting a survey of viruses in a hot acidic lake in Lassen Volcanic National Park, California, they uncovered evidence that a simple DNA virus had acquired a gene from a completely unrelated RNA-based virus. Virologist Luis Villareal of the University of California Irvine also suggests that viruses capable of converting an RNA-based gene into DNA and then incorporating it into a more complex DNA-based genome might have been common in the Virus world during the RNA to DNA transition some 4 billion years ago. This finding bolsters the argument for the transfer of information from the RNA world to the emerging DNA world before the emergence of the last universal common ancestor. From the research, the diversity of this virus world is still with us.
Viroids
Additional evidence supporting the concept of an RNA world has resulted from research on viroids, the first representatives of a novel domain of "subviral pathogens". Viroids are mostly plant pathogens, which consist of short stretches (a few hundred nucleobases) of highly complementary, circular, single-stranded, and non-coding RNA without a protein coat. Compared with other infectious plant pathogens, viroids are extremely small, ranging from 246 to 467 nucleobases. In comparison, the genome of the smallest known viruses capable of causing an infection are about 2,000 nucleobases long.
In 1989, Diener proposed that, based on their characteristic properties, viroids are more plausible "living relics" of the RNA world than are introns or other RNAs then so considered. If so, viroids have attained potential significance beyond plant pathology to evolutionary biology, by representing the most plausible macromolecules known capable of explaining crucial intermediate steps in the evolution of life from inanimate matter.
Apparently, Diener's hypothesis lay dormant until 2014, when Flores et al. published a review paper, in which Diener's evidence supporting his hypothesis was summarized. In the same year, a New York Times science writer published a popularized version of Diener's proposal, in which, however, he mistakenly credited Flores et al. with the hypothesis' original conception.
Pertinent viroid properties listed in 1989 are:
- small size, imposed by error-prone replication;
- high guanine and cytosine content, which increases stability and replication fidelity;
- circular structure, which assures complete replication without genomic tags;
- structural periodicity, which permits modular assembly into enlarged genomes;
- lack of protein-coding ability, consistent with a ribosome-free habitat; and
- in some cases, replication mediated by ribozymes—the fingerprint of the RNA world.
The existence, in extant cells, of RNAs with molecular properties predicted for RNAs of the RNA World constitutes an additional argument supporting the RNA World hypothesis.
Origin of sexual reproduction
Eigen et al. and Woese proposed that the genomes of early protocells were composed of single-stranded RNA, and that individual genes corresponded to separate RNA segments, rather than being linked end-to-end as in present-day DNA genomes. A protocell that was haploid (one copy of each RNA gene) would be vulnerable to damage, since a single lesion in any RNA segment would be potentially lethal to the protocell (e.g. by blocking replication or inhibiting the function of an essential gene).
Vulnerability to damage could be reduced by maintaining two or more copies of each RNA segment in each protocell, i.e. by maintaining diploidy or polyploidy. Genome redundancy would allow a damaged RNA segment to be replaced by an additional replication of its homolog. However, for such a simple organism, the proportion of available resources tied up in the genetic material would be a large fraction of the total resource budget. Under limited resource conditions, the protocell reproductive rate would likely be inversely related to ploidy number. The protocell's fitness would be reduced by the costs of redundancy. Consequently, coping with damaged RNA genes while minimizing the costs of redundancy would likely have been a fundamental problem for early protocells.
A cost-benefit analysis was carried out in which the costs of maintaining redundancy were balanced against the costs of genome damage. This analysis led to the conclusion that, under a wide range of circumstances, the selected strategy would be for each protocell to be haploid, but to periodically fuse with another haploid protocell to form a transient diploid. The retention of the haploid state maximizes the growth rate. The periodic fusions permit mutual reactivation of otherwise lethally damaged protocells. If at least one damage-free copy of each RNA gene is present in the transient diploid, viable progeny can be formed. For two, rather than one, viable daughter cells to be produced would require an extra replication of the intact RNA gene homologous to any RNA gene that had been damaged prior to the division of the fused protocell. The cycle of haploid reproduction, with occasional fusion to a transient diploid state, followed by splitting to the haploid state, can be considered to be the sexual cycle in its most primitive form. In the absence of this sexual cycle, haploid protocells with damage in an essential RNA gene would simply die.
This model for the early sexual cycle is hypothetical, but it is very similar to the known sexual behavior of the segmented RNA viruses, which are among the simplest organisms known. Influenza virus, whose genome consists of 8 physically separated single-stranded RNA segments, is an example of this type of virus. In segmented RNA viruses, "mating" can occur when a host cell is infected by at least two virus particles. If these viruses each contain an RNA segment with a lethal damage, multiple infection can lead to reactivation providing that at least one undamaged copy of each virus gene is present in the infected cell. This phenomenon is known as "multiplicity reactivation". Multiplicity reactivation has been reported to occur in influenza virus infections after induction of RNA damage by UV-irradiation, and ionizing radiation.
Further developments
Patrick Forterre has been working on a novel hypothesis, called "three viruses, three domains": that viruses were instrumental in the transition from RNA to DNA and the evolution of Bacteria, Archaea, and Eukaryota. He believes the last universal common ancestor was RNA-based and evolved RNA viruses. Some of the viruses evolved into DNA viruses to protect their genes from attack. Through the process of viral infection into hosts the three domains of life evolved.
Another interesting proposal is the idea that RNA synthesis might have been driven by temperature gradients, in the process of thermosynthesis. Single nucleotides have been shown to catalyze organic reactions.
Steven Benner has argued that chemical conditions on the planet Mars, such as the presence of boron, molybdenum, and oxygen, may have been better for initially producing RNA molecules than those on Earth. If so, life-suitable molecules, originating on Mars, may have later migrated to Earth via mechanisms of panspermia or similar process.
Alternative hypotheses
The hypothesized existence of an RNA world does not exclude a "Pre-RNA world", where a metabolic system based on a different nucleic acid is proposed to pre-date RNA. A candidate nucleic acid is peptide nucleic acid (PNA), which uses simple peptide bonds to link nucleobases. PNA is more stable than RNA, but its ability to be generated under prebiological conditions has yet to be demonstrated experimentally.
Threose nucleic acid (TNA) has also been proposed as a starting point, as has glycol nucleic acid (GNA), and like PNA, also lack experimental evidence for their respective abiogenesis.
An alternative—or complementary—theory of RNA origin is proposed in the PAH world hypothesis, whereby polycyclic aromatic hydrocarbons (PAHs) mediate the synthesis of RNA molecules. PAHs are the most common and abundant of the known polyatomic molecules in the visible Universe, and are a likely constituent of the primordial sea. PAHs and fullerenes (also implicated in the origin of life) have been detected in nebulae.
The iron-sulfur world theory proposes that simple metabolic processes developed before genetic materials did, and these energy-producing cycles catalyzed the production of genes.
Some of the difficulties of producing the precursors on earth are bypassed by another alternative or complementary theory for their origin, panspermia. It discusses the possibility that the earliest life on this planet was carried here from somewhere else in the galaxy, possibly on meteorites similar to the Murchison meteorite. sugar molecules, including ribose, have been found in meteorites. Panspermia does not invalidate the concept of an RNA world, but posits that this world or its precursors originated not on Earth but rather another, probably older, planet.
There are hypotheses that are in direct conflict to the RNA world hypothesis. The relative chemical complexity of the nucleotide and the unlikelihood of it spontaneously arising, along with the limited number of combinations possible among four base forms, as well as the need for RNA polymers of some length before seeing enzymatic activity, have led some to reject the RNA world hypothesis in favor of a metabolism-first hypothesis, where the chemistry underlying cellular function arose first, along with the ability to replicate and facilitate this metabolism.
RNA-peptide coevolution
Another proposal is that the dual-molecule system we see today, where a nucleotide-based molecule is needed to synthesize protein, and a peptide-based (protein) molecule is needed to make nucleic acid polymers, represents the original form of life. This theory is called RNA-peptide coevolution, or the Peptide-RNA world, and offers a possible explanation for the rapid evolution of high-quality replication in RNA (since proteins are catalysts), with the disadvantage of having to postulate the coincident formation of two complex molecules, an enzyme (from peptides) and a RNA (from nucleotides). In this Peptide-RNA World scenario, RNA would have contained the instructions for life, while peptides (simple protein enzymes) would have accelerated key chemical reactions to carry out those instructions. The study leaves open the question of exactly how those primitive systems managed to replicate themselves — something neither the RNA World hypothesis nor the Peptide-RNA World theory can yet explain, unless polymerases (enzymes that rapidly assemble the RNA molecule) played a role.
A research project completed in March 2015 by the Sutherland group found that a network of reactions beginning with hydrogen cyanide and hydrogen sulfide, in streams of water irradiated by UV light, could produce the chemical components of proteins and lipids, alongside those of RNA. The researchers used the term "cyanosulfidic" to describe this network of reactions. In November 2017, a team at the Scripps Research Institute identified reactions involving the compound diamidophosphate which could have linked the chemical components into short peptide and lipid chains as well as short RNA-like chains of nucleotides.
Implications
The RNA world hypothesis, if true, has important implications for the definition of life. For most of the time that followed Watson and Crick's elucidation of DNA structure in 1953, life was largely defined in terms of DNA and proteins: DNA and proteins seemed the dominant macromolecules in the living cell, with RNA only aiding in creating proteins from the DNA blueprint.
The RNA world hypothesis places RNA at center-stage when life originated. The RNA world hypothesis is supported by the observations that ribosomes are ribozymes: the catalytic site is composed of RNA, and proteins hold no major structural role and are of peripheral functional importance. This was confirmed with the deciphering of the 3-dimensional structure of the ribosome in 2001. Specifically, peptide bond formation, the reaction that binds amino acids together into proteins, is now known to be catalyzed by an adenine residue in the rRNA.
RNAs are known to play roles in other cellular catalytic processes, specifically in the targeting of enzymes to specific RNA sequences. In eukaryotes, the processing of pre-mRNA and RNA editing take place at sites determined by the base pairing between the target RNA and RNA constituents of small nuclear ribonucleoproteins (snRNPs). Such enzyme targeting is also responsible for gene down regulation though RNA interference (RNAi), where an enzyme-associated guide RNA targets specific mRNA for selective destruction. Likewise, in eukaryotes the maintenance of telomeres involves copying of an RNA template that is a constituent part of the telomerase ribonucleoprotein enzyme. Another cellular organelle, the vault, includes a ribonucleoprotein component, although the function of this organelle remains to be elucidated.
Interestingly, the "Alanine World" hypothesis places the canonical amino acid Alanine in the centre of the so-called Protein-World. Dominant secondary structures in modern proteins are α-helices and β-sheets. The most commonly selected monomers (i.e. amino acids) for ribosomal protein synthesis are chemical derivatives of the α-amino acid Alanine as they are best suited for the construction of α-helices or β-sheets in modern proteins.