From Wikipedia, the free encyclopedia
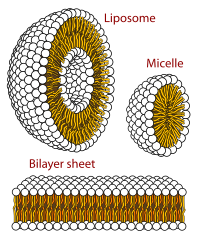
The three main structures phospholipids form in solution; the liposome (a closed bilayer), the micelle and the bilayer.
A protocell (or protobiont) is a self-organized, endogenously ordered, spherical collection of lipids proposed as a stepping-stone to the origin of life.[1][2] A central question in evolution is how simple protocells first arose and how they could differ in reproductive output, thus enabling the accumulation of novel biological emergences over time, i.e. biological evolution. Although a functional protocell has not yet been achieved in a laboratory setting, the goal to understand the process appears well within reach.[3][4][5][6]
Overview
Compartmentalization was important in the origins of life. Membranes create enclosed compartments that are separate from the external environment, thus providing the cell with functionally specialized aqueous spaces.Because lipid bilayer of membranes is impermeable to most hydrophilic molecules (dissolved by water), the cell must have membrane transport systems that are in charge of import of nutritive molecules as well as export of waste.[7] It is very challenging to construct protocells from molecular assemblies. An important step in this challenge is the achievement of vesicle dynamics that are relevant to cellular functions, such as membrane trafficking and self-reproduction, using amphiphilic molecules. On the primitive Earth, numerous chemical reactions of organic compounds produced the ingredients of life. Of these substances, amphiphilic molecules might be the first player in the evolution from molecular assembly to cellular life.[8][9] A step from vesicle toward protocell might be to develop self-reproducing vesicles coupled with the metabolic system.[10]
Selectivity for compartmentalization
Self-assembled vesicles are essential components of primitive cells.[1] The second law of thermodynamics requires that the universe move in a direction in which disorder (or entropy) increases, yet life is distinguished by its great degree of organization. Therefore, a boundary is needed to separate life processes from non-living matter.[11] The cell membrane is the only cellular structure that is found in all of the cells of all of the organisms on Earth.[12]Researchers Irene A. Chen and Jack W. Szostak (Nobel Prize in Physiology or Medicine 2009) amongst others, demonstrated that simple physicochemical properties of elementary protocells can give rise to essential cellular behaviors, including primitive forms of Darwinian competition and energy storage. Such cooperative interactions between the membrane and encapsulated contents could greatly simplify the transition from replicating molecules to true cells.[4] Furthermore, competition for membrane molecules would favor stabilized membranes, suggesting a selective advantage for the evolution of cross-linked fatty acids and even the phospholipids of today.[4] This micro-encapsulation allowed for metabolism within the membrane, exchange of small molecules and prevention of passage of large substances across it.[13] The main advantages of encapsulation include increased solubility of the cargo and creating energy in the form of chemical gradient. Energy is thus often said to be stored by cells in the structures of molecules of substances such as carbohydrates (including sugars), lipids, and proteins, which release energy when chemically combined with oxygen during cellular respiration.[14][15]
Energy gradient
A March 2014 study by NASA's Jet Propulsion Laboratory, demonstrated a unique way to study the origins of life: fuel cells.[16] Fuel cells are similar to biological cells in that electrons are also transferred to and from molecules. In both cases, this results in electricity and power. The study states that one important factor was that the Earth provides electrical energy at the seafloor. "This energy could have kick-started life and could have sustained life after it arose. Now, we have a way of testing different materials and environments that could have helped life arise not just on Earth, but possibly on Mars, Europa and other places in the Solar System."[16]Vesicles and micelles
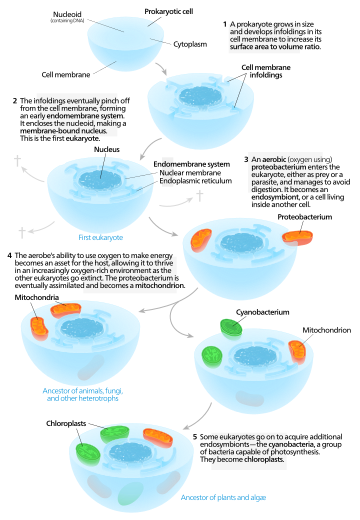
When phospholipids are placed in water, the molecules spontaneously arrange such that the tails are shielded from the water, resulting in the formation of membrane structures such as bilayers, vesicles, and micelles.[2] In modern cells, vesicles are involved in metabolism, transport, buoyancy control,[17] and enzyme storage. They can also act as natural chemical reaction chambers. A typical vesicle or micelle in aqueous solution forms an aggregate with the hydrophilic "head" regions in contact with surrounding solvent, sequestering the hydrophobic single-tail regions in the micelle centre. This phase is caused by the packing behavior of single-tail lipids in a bilayer. Although the protocellular self-assembly process that spontaneously form lipid monolayer vesicles and micelles in nature resemble the kinds of primordial vesicles or protocells that might have existed at the beginning of evolution, they are not as sophisticated as the bilayer membranes of today's living organisms.[18]
Rather than being made up of phospholipids, however, early membranes may have formed from monolayers or bilayers of fatty acids, which may have formed more readily in a prebiotic environment.[19] Fatty acids have been synthesized in laboratories under a variety of prebiotic conditions and have been found on meteorites, suggesting their natural synthesis in nature.[4]
Oleic acid vesicles represent good models of membrane protocells that could have existed in prebiotic times.[20]
Geothermal ponds and clay
Scientists have come to conclude that life began in hydrothermal vents in the deep sea, but a 2012 study led by Armen Mulkidjanian of Germany's University of Osnabrück, suggests that inland pools of condensed and cooled geothermal vapor have the ideal characteristics for the origin of life.[21] The conclusion is based mainly on the chemistry of modern cells, where the cytoplasm is rich in potassium, zinc, manganese, and phosphate ions, which are not widespread in marine environments. Such conditions, the researchers argue, are found only where hot hydrothermal fluid brings the ions to the surface — places such as geysers, mud pots, fumaroles and other geothermal features. Within these fuming and bubbling basins, water laden with zinc and manganese ions could have collected, cooled and condensed in shallow pools.[21]
In the 1990s biochemist James Ferris of Rensselaer Polytechnic Institute showed that montmorillonite clay can help create RNA chains of as many as 50 nucleotides joined together spontaneously into a single RNA molecule.[5] Then in 2002, Hanczyc, Fujikawa and Szostak discovered that by adding montmorillonite to their solution of fatty acid micelles (lipid spheres), the clay sped up the rate of vesicles formation 100-fold.[5]
Research has shown that some minerals can catalyze the stepwise formation of hydrocarbon tails of fatty acids from hydrogen and carbon monoxide gases - gases that may have been released from hydrothermal vents or geysers.
Fatty acids of various lengths are eventually released into the surrounding water,[19] but vesicle formation requires a higher concentration of fatty acids, so it is suggested that protocell formation started at land-bound hydrothermal vents such as geysers, mud pots, fumaroles and other geothermal features where water evaporates and concentrates the solute.[5][22][23]
Montmorillonite bubbles
A team of applied physicists at Harvard's School of Engineering and Applied Sciences say that primitive cells might have formed inside inorganic clay microcompartments, which can provide an ideal container for the synthesis and compartmentalization of complex organic molecules.[24] Clay-armored "bubbles" form naturally when particles of montmorillonite clay collect on the outer surface of air bubbles under water. This creates a semi permeable vesicle from materials that are readily available in the environment. The authors remark that montmorillonite is known to serve as a chemical catalyst, encouraging lipids to form membranes and single nucleotides to join into strands of RNA. Primitive reproduction can be envisioned when the clay bubbles burst, releasing the lipid membrane-bound product into the surrounding medium.[24]Membrane transport
Instead of the more popular phospholipids of modern cells, the membrane of protocells in the RNA world would be composed of fatty acids,[25] and that such membranes have relatively high permeability to ions and small molecules,[1] such as nucleoside monophosphate (NMP), nucleoside diphosphate (NDP), and nucleoside triphosphatee (NTP), and may withstand millimolar concentrations of Mg2+.[26] Osmotic pressure also plays a significant role in protocell membrane transport.[1]
It has been proposed that electroporation resulting from lightning strikes could be a mechanism of natural horizontal gene transfer.[27] Electroporation is the rapid increase in bilayer permeability induced by the application of a large artificial electric field across the membrane. During electroporation in laboratory procedures, the lipid molecules are not chemically altered but simply shift position, opening up a pore (hole) that acts as the conductive pathway through the bilayer as it is filled with water. The mechanism is the creation of nanometer sized water-filled holes in the membrane. Experimentally, electroporation is used to introduce hydrophilic molecules into cells. It is a particularly useful technique for large highly charged molecules such as DNA and RNA, which would never passively diffuse across the hydrophobic bilayer core.[28] Because of this, electroporation is one of the key methods of transfection as well as bacterial transformation.
- Fusion
Artificial models
Langmuir-Blodgett deposition
Starting with a technique commonly used to deposit molecules on a solid surface, Langmuir-Blodgett deposition, scientist are able to assemble phospholipid membranes layer by layer of arbitrary complexity.[30][31] These artificial phospholipid membranes support functional insertion both of purified and of in situ expressed membrane proteins.[31] The technique could help astrobiologists understand how the first living cells originated.[30]
Jeewanu
The Jeewanu protocells are synthetic chemical particles that possess cell-like structure and seem to have some functional living properties.[32] First synthesized in 1963 from simple minerals and basic organics while exposed to sunlight, it is still reported to have some metabolic capabilities, the presence of semipermeable membrane, amino acids, phospholipids, carbohydrates and RNA-like molecules.[32][33] However, the nature and properties of the Jeewanu remains to be clarified.[32][33][34]In a similar synthesis experiment using light, led by Jason Dworkin in 2000,[35] he exposed a frozen mixture of water, methanol, ammonia and carbon monoxide to ultraviolet (UV) radiation. This combination yielded large amounts of organic material that self-organised to form globules or vesicles when immersed in water. Dworkin considered these globules to resemble cell membranes that enclose and concentrate the chemistry of life, separating their interior from the outside world. The globules were between 10 to 40 micrometres (0.00039 to 0.00157 in), or about the size of red blood cells. Remarkably, the globules fluoresced, or glowed, when exposed to UV light.
Absorbing UV and converting it into visible light in this way was considered one possible way of providing energy to a primitive cell. If such globules played a role in the origin of life, the fluorescence could have been a precursor to primitive photosynthesis. Such fluorescence also provides the benefit of acting as a sunscreen, diffusing any damage that otherwise would be inflicted by UV radiation. Such a protective function would have been vital for life on the early Earth, since the ozone layer, which blocks out the sun's most destructive UV rays, did not form until after photosynthetic life began to produce oxygen.[36]