| Cascade (CRISPR-associated complex for antiviral defense) | |
|---|---|
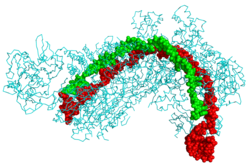
CRISPR Cascade protein (cyan) bound to CRISPR RNA (green) and viral DNA (red).
|
|
| Identifiers | |
| Organism | |
| Symbol | CRISPR |
| PDB | 4QYZ |
CRISPR (/ˈkrɪspər/) is a family of DNA sequences in bacteria and archaea.[1] The sequences contain snippets of DNA from viruses that have attacked the prokaryote. These snippets are used by the prokaryote to detect and destroy DNA from similar viruses during subsequent attacks. These sequences play a key role in a prokaryotic defense system,[1] and form the basis of a technology known as CRISPR/Cas9 that effectively and specifically changes genes within organisms.[2]
CRISPR is an abbreviation of Clustered Regularly Interspaced Short Palindromic Repeats.[3] The name was minted at a time when the origin and use of the interspacing subsequences were not known. At that time the CRISPRs were described as segments of prokaryotic DNA containing short, repetitive base sequences. In a palindromic repeat, the sequence of nucleotides is the same in both directions. Each repetition is followed by short segments of spacer DNA from previous exposures to foreign DNA (e.g., a virus or plasmid).[4][5] Small clusters of cas (CRISPR-associated system) genes are located next to CRISPR sequences.
CRISPR/Cas9
The CRISPR/Cas system is a prokaryotic immune system that confers resistance to foreign genetic elements such as those present within plasmids and phages[6][7][8] that provides a form of acquired immunity. RNA harboring the spacer sequence helps Cas (CRISPR-associated) proteins recognize and cut exogenous DNA. Other RNA-guided Cas proteins cut foreign RNA.[9] CRISPRs are found in approximately 50% of sequenced bacterial genomes and nearly 90% of sequenced archaea.[10]
Use for gene editing
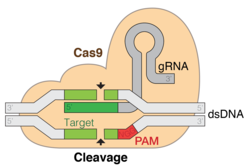
A simple version of the CRISPR/Cas system, CRISPR/Cas9, has been modified to edit genomes. By delivering the Cas9 nuclease complexed with a synthetic guide RNA (gRNA) into a cell, the cell's genome can be cut at a desired location, allowing existing genes to be removed and/or new ones added.[11][12][13] The Cas9-gRNA complex corresponds with the CAS III CRISPR-RNA complex in the above diagram.
Diagram of the CRISPR prokaryotic antiviral defense mechanism.[14]
CRISPR/Cas genome editing techniques have many potential applications, including medicine and crop seed enhancement. The use of CRISPR/Cas9-gRNA complex for genome editing[15][16] was the AAAS's choice for breakthrough of the year in 2015.[17] Bioethical concerns have been raised about the prospect of using CRISPR for germline editing.[18]
History
Repeated sequences
The discovery of clustered DNA repeats occurred independently in three parts of the world. The first description of what would later be called CRISPR is from Osaka University researcher Yoshizumi Ishino and his colleagues in 1987. They accidentally cloned part of a CRISPR together with the iap gene, the target of interest. The organization of the repeats was unusual because repeated sequences are typically arranged consecutively along DNA.[19][20] They studied the relation of "iap" to the bacterium E. coli. The function of the interrupted clustered repeats was not known at the time.In 1993 researchers of Mycobacterium tuberculosis in the Netherlands published two articles about a cluster of interrupted direct repeats (DR) in this bacterium. These researchers recognized the diversity of the DR-intervening sequences among different strains of M. tuberculosis[21] and used this property to design a typing method that was named spoligotyping, which is still in use today.[22][23]
At the same time, repeats were observed in the archaeal organisms of Haloferax and Haloarcula species, and their function was studied by Francisco Mojica at the University of Alicante in Spain. Although his hypothesis turned out to be wrong, Mojica's supervisor surmised at the time that the clustered repeats had a role in correctly segregating replicated DNA into daughter cells during cell division because plasmids and chromosomes with identical repeat arrays could not coexist in Haloferax volcanii. Transcription of the interrupted repeats was also noted for the first time.[23][24] By 2000, Mojica performed a survey of scientific literature and one of his students a search in published genomes with program devised by himself. They identified interrupted repeats in 20 species of microbes as belonging to the same family.[5] In 2001, Mojica and Ruud Jansen, who was searching for additional interrupted repeats, proposed the acronym CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) to alleviate the confusion stemming from the numerous acronyms used to describe the sequences in the scientific literature.[24][25]
CRISPR-associated systems
A major addition to the understanding of CRISPR came with Jansen's observation that the prokaryote repeat cluster was accompanied by a set of homologous genes that make up CRISPR-associated systems or cas genes. Four cas genes (cas 1 - 4) were initially recognized. The Cas proteins showed helicase and nuclease motifs, suggesting a role in the dynamic structure of the CRISPR loci.[26] In this publication the acronym CRISPR was coined as the universal name of this pattern. However, the CRISPR function remained enigmatic.
Simplified diagram of a CRISPR locus. The three major components of a CRISPR locus are shown: cas
genes, a leader sequence, and a repeat-spacer array. Repeats are shown
as gray boxes and spacers are colored bars. The arrangement of the three
components is not always as shown.[14][4] In addition, several CRISPRs with similar sequences can be present in a single genome, only one of which is associated with cas genes.[27]
In 2005, three independent research groups showed that some CRISPR spacers are derived from phage DNA and extrachromosomal DNA such as plasmids.[28][29][30] In effect, the spacers are fragments of DNA gathered from viruses that previously tried to attack the cell. The source of the spacers was a sign that the CRISPR/cas system could have a role in adaptive immunity in bacteria.[14][31] All three studies proposing this idea were initially rejected by high-profile journals, but eventually appeared in other journals.[32]
The first publication[29] proposing a role of CRISPR-Cas in microbial immunity, by the researchers at the University of Alicante, predicted a role for the RNA transcript of spacers on target recognition in a mechanism that could be analogous to the RNA interference system used by eukaryotic cells. Koonin and colleagues extended this RNA interference hypothesis by proposing mechanisms of action for the different CRISPR-Cas subtypes according to the predicted function of their proteins.[33]
Experimental work by several groups revealed the basic mechanisms of CRISPR-Cas immunity. In 2007 the first experimental evidence that CRISPR was an adaptive immune system was published.[20] A CRISPR region in Streptococcus thermophilus acquired spacers from the DNA of an infecting bacteriophage. The researchers manipulated the resistance of S. thermophilus to phage by adding and deleting spacers whose sequence matched those found in the tested phages.[34][35] In 2008, Brouns and Van der Oost identified a complex of Cas proteins (called Cascade) that in E. coli cut the CRISPR RNA precursor within the repeats into mature spacer-containing RNA molecules (crRNA), which remained bound to the protein complex.[36] Moreover, it was found that Cascade, crRNA and an helicase/nuclease (Cas3) were required to provide a bacterial host with immunity against infection by a DNA virus. By designing an anti-virus CRISPR, they demonstrated that two orientations of the crRNA (sense/antisense) provided immunity, indicating that the crRNA guides were targeting dsDNA. That year Marraffini and Sontheimer indeed confirmed that a CRISPR sequence of S. epidermidis targeted DNA and not RNA to prevent conjugation. This finding was at odds with the proposed RNA-interference-like mechanism of CRISPR-Cas immunity, although a CRISPR-Cas system that targets foreign RNA was later found in Pyrococcus furiosus.[20][34] A 2010 study showed that CRISPR-Cas cuts both strands of phage and plasmid DNA in S. thermophilus.[37]
Cas9
Researchers studied a simpler CRISPR system from Streptococcus pyogenes that relies on the protein Cas9. The Cas9 endonuclease is a four-component system that includes two small RNA molecules named CRISPR RNA (crRNA) and trans-activating CRISPR RNA (tracrRNA).[38] Jennifer Doudna and Emmanuelle Charpentier re-engineered the Cas9 endonuclease into a more manageable two-component system by fusing the two RNA molecules into a "single-guide RNA" that, when combined with Cas9, could find and cut the DNA target specified by the guide RNA. By manipulating the nucleotide sequence of the guide RNA, the artificial Cas9 system could be programmed to target any DNA sequence for cleavage.[39] Another group of collaborators comprising Šikšnys together with Gasiūnas, Barrangou and Horvath showed that Cas9 from the S. thermophilus CRISPR system can also be reprogrammed to target a site of their choosing by changing the sequence of its crRNA. These advances fueled efforts to edit genomes with the modified CRISPR-Cas9 system.[23]Feng Zhang's and George Church's groups simultaneously described genome editing in human cell cultures using CRISPR-Cas9 for the first time.[20][40][41] It has since been used in a wide range of organisms, including baker's yeast (Saccharomyces cerevisiae),[42][43][44] the opportunistic pathogen C. albicans,[45][46] zebrafish (D. rerio),[47] fruit flies (Drosophila melanogaster),[48] nematodes (C. elegans),[49] plants,[50] mice,[51] monkeys[52] and human embryos.[53]
CRISPR has been modified to make programmable transcription factors that allow scientists to target and activate or silence specific genes.[54]
The CRIPSR/Cas9 system has shown to make effective gene edits in Human tripronuclear zygotes first described in a 2015 paper by Chinese scientists P. Liang and Y. Xu. The system made a successful cleavage of mutant Beta-Hemoglobin (HBB) in 28 out of 54 embryos. 4 out of the 28 embryos were successfully recombined using a donor template given by the scientists. The scientists showed that during DNA recombination of the cleaved strand, the homologous endogenous sequence HBD competes with the exogenous donor template. DNA repair in human embryos is much more complicated and particular than in derived stem cells.[55]
Cpf1
In 2015, the nuclease Cpf1 was discovered in the CRISPR/Cpf1 system of the bacterium Francisella novicida.[56][57] Cpf1 showed several key differences from Cas9 including: causing a 'staggered' cut in double stranded DNA as opposed to the 'blunt' cut produced by Cas9, relying on a 'T rich' PAM (providing alternate targeting sites to Cas9) and requiring only a CRISPR RNA (crRNA) for successful targeting. By contrast Cas9 requires both crRNA and a transactivating crRNA (tracrRNA).These differences may give Cpf1 some advantages over Cas9. For example, Cpf1's small crRNAs are ideal for multiplexed genome editing, as more of them can be packaged in one vector than can Cas9's sgRNAs. As well, the sticky 5' overhangs left by Cpf1 can be used for DNA assembly that is much more target-specific than traditional Restriction Enzyme cloning.[58] Finally, Cpf1 cleaves DNA 18-23 bp downstream from the PAM site. This means there is no disruption to the recognition sequence after repair, and so Cpf1 enables multiple rounds of DNA cleavage. By contrast, since Cas9 cuts only 3 bp upstream of the PAM site, the NHEJ pathway results in indel mutations which destroy the recognition sequence, thereby preventing further rounds of cutting. In theory, repeated rounds of DNA cleavage should cause an increased opportunity for the desired genomic editing to occur.[59]
Locus structure
Repeats and spacers
The CRISPR array comprises an AT-rich leader sequence followed by short repeats that are separated by unique spacers.[60] CRISPR repeats typically range in size from 28 to 37 base pairs (bps), though there can be as few as 23 bp and as many as 55 bp.[61] Some show dyad symmetry, implying the formation of a secondary structure such as a stem-loop ('hairpin') in the RNA, while others are predicted to be unstructured. The size of spacers in different CRISPR arrays is typically 32 to 38 bp (range 21 to 72 bp).[61] New spacers can appear rapidly as part of the immune response to phage infection.[62] There are usually fewer than 50 units of the repeat-spacer sequence in a CRISPR array.[61]Cas genes and CRISPR subtypes
Small clusters of cas genes are often located next to CRISPR repeat-spacer arrays. Collectively there are 93 cas genes that are grouped into 35 families based on sequence similarity of the encoded proteins. 11 of the 35 families form the cas core, which includes the protein families Cas1 through Cas9. A complete CRISPR-Cas locus has at least one gene belonging to the cas core.[63]CRISPR-Cas systems fall into two classes. Class 1 systems use a complex of multiple Cas proteins to degrade foreign nucleic acids. Class 2 systems use a single large Cas protein for the same purpose. Class 1 is divided into types I, III, and IV; class 2 is divided into types II, V, and VI.[64] The 6 system types are divided into 19 subtypes.[65] Each type and most subtypes are characterized by a "signature gene" found almost exclusively in the category. Classification is also based on the complement of cas genes that are present. Most CRISPR-Cas systems have a Cas1 protein. The phylogeny of Cas1 proteins generally agrees with the classification system.[63] Many organisms contain multiple CRISPR-Cas systems suggesting that they are compatible and may share components.[66][67] The sporadic distribution of the CRISPR/Cas subtypes suggests that the CRISPR/Cas system is subject to horizontal gene transfer during microbial evolution.
| Class | Cas type | Signature protein | Function | Reference |
|---|---|---|---|---|
| 1 | I | Cas3 | Single-stranded DNA nuclease (HD domain) and ATP-dependent helicase | [68][69] |
| IA | Cas8a, Cas5 | Subunit of the interference module. Important in targeting of invading DNA by recognizing the PAM sequence | [63] | |
| IB | Cas8b | |||
| IC | Cas8c | |||
| ID | Cas10d | contains a domain homologous to the palm domain of nucleic acid polymerases and nucleotide cyclases | [70][71] | |
| IE | Cse1, Cse2 | |||
| IF | Csy1, Csy2, Csy3 | Not determined | [63] | |
| IU | GSU0054 | [63] | ||
| III | Cas10 | Homolog of Cas10d and Cse1 | [71] | |
| IIIA | Csm2 | Not Determined | [63] | |
| IIIB | Cmr5 | Not Determined | [63] | |
| IIIC | Cas10 or Csx11 | [63] | ||
| IIID | Csx10 | [63] | ||
| IV | Csf1 | |||
| IVA | ||||
| IVB | ||||
| 2 | II | Cas9 | Nucleases RuvC and HNH together produce DSBs, and separately can produce single-strand breaks. Ensures the acquisition of functional spacers during adaptation. | [72][73] |
| IIA | Csn2 | Ring-shaped DNA-binding protein. Involved in primed adaptation in Type II CRISPR system. | [74] | |
| IIB | Cas4 | Not Determined | ||
| IIC | Characterized by the absence of either Csn2 or Cas4 | [75] | ||
| V | Cpf1, C2c1, C2c3 | Nuclease RuvC. Lacks HNH. | [64] | |
| VI | Cas13a (previously known as C2c2), Cas13b, Cas13c | RNA-guided RNase | [64] [76] |
Mechanism
The CRISPR genetic locus provides bacteria with a defense mechanism to protect them from repeated phage infections.
Transcripts of the CRISPR Genetic Locus and Maturation of pre-crRNA
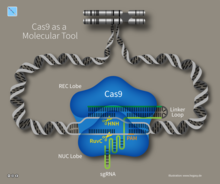
3D Structure of the CRISPR-Cas9 Interference Complex
CRISPR-Cas9 as a Molecular Tool Introduces Targeted Double Strand DNA Breaks.
Double Strand DNA Breaks Introduced by CRISPR-Cas9 Allows Further
Genetic Manipulation By Exploiting Endogenous DNA Repair Mechanisms.

The stages of CRISPR immunity for each of the
three major types of adaptive immunity. (1) Acquisition begins by
recognition of invading DNA by Cas1
and Cas2 and cleavage of a protospacer. (2) The protospacer is ligated
to the direct repeat adjacent to the leader sequence and (3) single
strand extension repairs the CRISPR and duplicates the direct repeat.
The crRNA processing and interference stages occur differently in each
of the three major CRISPR systems. (4) The primary CRISPR transcript is
cleaved by cas genes to produce crRNAs. (5) In type I systems
Cas6e/Cas6f cleave at the junction of ssRNA and dsRNA formed by hairpin
loops in the direct repeat. Type II systems use a trans-activating
(tracr) RNA to form dsRNA, which is cleaved by Cas9
and RNaseIII. Type III systems use a Cas6 homolog that does not require
hairpin loops in the direct repeat for cleavage. (6) In type II and
type III systems secondary trimming is performed at either the 5’ or 3’
end to produce mature crRNAs. (7) Mature crRNAs associate with Cas
proteins to form interference complexes. (8) In type I and type II
systems, interactions between the protein and PAM sequence are required
for degradation of invading DNA. Type III systems do not require a PAM
for successful degradation and in type III-A systems basepairing occurs
between the crRNA and mRNA rather than the DNA, targeted by type III-B
systems.
CRISPR-Cas immunity is a natural process of bacteria and archaea. CRISPR-Cas prevents bacteriophage infection, conjugation and natural transformation by degrading foreign nucleic acids that enter the cell.[34]
Spacer acquisition
When a microbe is invaded by a virus, the first stage of the immune response is to capture viral DNA and insert it into a CRISPR locus in the form of a spacer. Cas1 and Cas2 are found in all two types of CRISPR-Cas immune systems, which indicates that they are involved in spacer acquisition. Mutation studies confirmed this hypothesis, showing that removal of cas1 or cas2 stopped spacer acquisition, without affecting CRISPR immune response.[77][78][79][80][81]Multiple Cas1 proteins have been characterised and their structures resolved.[82][83][84] Cas1 proteins have diverse amino acid sequences. However, their crystal structures are similar and all purified Cas1 proteins are metal-dependent nucleases/integrases that bind to DNA in a sequence-independent manner.[66] Representative Cas2 proteins have been characterised and possess either (single strand) ssRNA-[85] or (double strand) dsDNA-[86][87] specific endoribonuclease activity.
In the I-E system of E. coli Cas1 and Cas2 form a complex where a Cas2 dimer bridges two Cas1 dimers.[88] In this complex Cas2 performs a non-enzymatic scaffolding role,[88] binding double-stranded fragments of invading DNA, while Cas1 binds the single-stranded flanks of the DNA and catalyses their integration into CRISPR arrays.[89][90][91] New spacers are usually added at the beginning of the CRISPR next to the leader sequence creating a chronological record of viral infections.[92] In E. Coli a histone like protein called integration host factor (IHF), which binds to the leader sequence, is responsible for the accuracy of this integration.[93]
Protospacer adjacent motifs
Bioinformatic analysis of regions of phage genomes that were excised as spacers (termed protospacers) revealed that they were not randomly selected but instead were found adjacent to short (3 – 5 bp) DNA sequences termed protospacer adjacent motifs (PAM). Analysis of CRISPR-Cas systems showed PAMs to be important for type I and type II, but not type III systems during acquisition.[30][94][95][96][97][98] In type I and type II systems, protospacers are excised at positions adjacent to a PAM sequence, with the other end of the spacer cut using a ruler mechanism, thus maintaining the regularity of the spacer size in the CRISPR array.[99][100] The conservation of the PAM sequence differs between CRISPR-Cas systems and appears to be evolutionarily linked to Cas1 and the leader sequence.[98][101]New spacers are added to a CRISPR array in a directional manner,[28] occurring preferentially,[62][94][95][102][103] but not exclusively, adjacent[97][100] to the leader sequence. Analysis of the type I-E system from E. coli demonstrated that the first direct repeat adjacent to the leader sequence, is copied, with the newly acquired spacer inserted between the first and second direct repeats.[80][99]
The PAM sequence appears to be important during spacer insertion in type I-E systems. That sequence contains a strongly conserved final nucleotide (nt) adjacent to the first nt of the protospacer. This nt becomes the final base in the first direct repeat.[81][104][105] This suggests that the spacer acquisition machinery generates single-stranded overhangs in the second-to-last position of the direct repeat and in the PAM during spacer insertion. However, not all CRISPR-Cas systems appear to share this mechanism as PAMs in other organisms do not show the same level of conservation in the final position.[101] It is likely that in those systems, a blunt end is generated at the very end of the direct repeat and the protospacer during acquisition.
Insertion variants
Analysis of Sulfolobus solfataricus CRISPRs revealed further complexities to the canonical model of spacer insertion, as one of its six CRISPR loci inserted new spacers randomly throughout its CRISPR array, as opposed to inserting closest to the leader sequence.[100]Multiple CRISPRs contain many spacers to the same phage. The mechanism that causes this phenomenon was discovered in the type I-E system of E. coli. A significant enhancement in spacer acquisition was detected where spacers already target the phage, even mismatches to the protospacer. This ‘priming’ requires the Cas proteins involved in both acquisition and interference to interact with each other. Newly acquired spacers that result from the priming mechanism are always found on the same strand as the priming spacer.[81][104][105] This observation led to the hypothesis that the acquisition machinery slides along the foreign DNA after priming to find a new protospacer.[105]
Biogenesis
CRISPR-RNA (crRNA), which later guides the Cas nuclease to the target during the interference step, must be generated from the CRISPR sequence. The crRNA is initially transcribed as part of a single long transcript encompassing much of the CRISPR array.[4] This transcript is then cleaved by Cas proteins to form crRNAs. The mechanism to produce crRNAs differs among CRISPR/Cas systems. In type I-E and type I-F systems, the proteins Cas6e and Cas6f respectively, recognise stem-loops[106][107][108] created by the pairing of identical repeats that flank the crRNA.[109] These Cas proteins cleave the longer transcript at the edge of the paired region, leaving a single crRNA along with a small remnant of the paired repeat region.Type III systems also use Cas6, however their repeats do not produce stem-loops. Cleavage instead occurs by the longer transcript wrapping around the Cas6 to allow cleavage just upstream of the repeat sequence.[110][111][112]
Type II systems lack the Cas6 gene and instead utilize RNaseIII for cleavage. Functional type II systems encode an extra small RNA that is complementary to the repeat sequence, known as a trans-activating crRNA (tracrRNA).[78] Transcription of the tracrRNA and the primary CRISPR transcript results in base pairing and the formation of dsRNA at the repeat sequence, which is subsequently targeted by RNaseIII to produce crRNAs. Unlike the other two systems the crRNA does not contain the full spacer, which is instead truncated at one end.[72]
CrRNAs associate with Cas proteins to form ribonucleotide complexes that recognize foreign nucleic acids. CrRNAs show no preference between the coding and non-coding strands, which is indicative of an RNA-guided DNA-targeting system.[8][37][77][81][113][114][115] The type I-E complex (commonly referred to as Cascade) requires five Cas proteins bound to a single crRNA.[116][117]
Interference
During the interference stage in type I systems the PAM sequence is recognized on the crRNA-complementary strand and is required along with crRNA annealing. In type I systems correct base pairing between the crRNA and the protospacer signals a conformational change in Cascade that recruits Cas3 for DNA degradation.Type II systems rely on a single multifunctional protein, Cas9, for the interference step.[72] Cas9 requires both the crRNA and the tracrRNA to function and cleaves DNA using its dual HNH and RuvC/RNaseH-like endonuclease domains. Basepairing between the PAM and the phage genome is required in type II systems. However, the PAM is recognized on the same strand as the crRNA (the opposite strand to type I systems).
Type III systems, like type I require six or seven Cas proteins binding to crRNAs.[118][119] The type III systems analysed from S. solfataricus and P. furiosus both target the mRNA of phages rather than phage DNA genome,[67][119] which may make these systems uniquely capable of targeting RNA-based phage genomes.[66]
The mechanism for distinguishing self from foreign DNA during interference is built into the crRNAs and is therefore likely common to all three systems. Throughout the distinctive maturation process of each major type, all crRNAs contain a spacer sequence and some portion of the repeat at one or both ends. It is the partial repeat sequence that prevents the CRISPR-Cas system from targeting the chromosome as base pairing beyond the spacer sequence signals self and prevents DNA cleavage.[120] RNA-guided CRISPR enzymes are classified as type V restriction enzymes.
| CRISPR associated protein | |||
|---|---|---|---|
crystal structure of a crispr-associated protein from Thermus thermophilus
|
|||
| Identifiers | |||
| Symbol | CRISPR_assoc | ||
| Pfam | PF08798 | ||
| Pfam clan | CL0362 | ||
| InterPro | IPR010179 | ||
| CDD | cd09727 | ||
| CRISPR associated protein Cas2 | |||
|---|---|---|---|

crystal structure of a hypothetical protein tt1823 from Thermus thermophilus
|
|||
| Identifiers | |||
| Symbol | CRISPR_Cas2 | ||
| Pfam | PF09827 | ||
| InterPro | IPR019199 | ||
| CDD | cd09638 | ||
|
|||
| CRISPR-associated protein Cse1 | |||
|---|---|---|---|
| Identifiers | |||
| Symbol | CRISPR_Cse1 | ||
| Pfam | PF09481 | ||
| InterPro | IPR013381 | ||
| CDD | cd09729 | ||
|
|||
| CRISPR-associated protein Cse2 | |||
|---|---|---|---|
| Identifiers | |||
| Symbol | CRISPR_Cse2 | ||
| Pfam | PF09485 | ||
| InterPro | IPR013382 | ||
| CDD | cd09670 | ||
|
|||
Evolution
A bioinformatic study has suggested that CRISPRs are evolutionarily conserved and cluster into related types. Many show signs of a conserved secondary structure.[109]CRISPR/Cas can immunize bacteria against certain phages and thus halt transmission. For this reason, Koonin described CRISPR/Cas as a Lamarckian inheritance mechanism.[121] However, this was disputed by a critic who noted, "We should remember [Lamarck] for the good he contributed to science, not for things that resemble his theory only superficially. Indeed, thinking of CRISPR and other phenomena as Lamarckian only obscures the simple and elegant way evolution really works".[122]
Coevolution
Analysis of CRISPR sequences revealed coevolution of host and viral genomes.[123] Cas9 proteins are highly enriched in pathogenic and commensal bacteria. CRISPR/Cas-mediated gene regulation may contribute to the regulation of endogenous bacterial genes, particularly during interaction with eukaryotic hosts. For example, Francisella novicida uses a unique, small, CRISPR/Cas-associated RNA (scaRNA) to repress an endogenous transcript encoding a bacterial lipoprotein that is critical for F. novicida to dampen host response and promote virulence.[124]The basic model of CRISPR evolution is newly incorporated spacers driving phages to mutate their genomes to avoid the bacterial immune response, creating diversity in both the phage and host populations. To fight off a phage infection, the sequence of the CRISPR spacer must correspond perfectly to the sequence of the target phage gene. Phages can continue to infect their hosts given point mutations in the spacer.[120] Similar stringency is required in PAM or the bacterial strain remains phage sensitive.[95][120]
Rates
A study of 124 S. thermophilus strains showed that 26% of all spacers were unique and that different CRISPR loci showed different rates of spacer acquisition.[94] Some CRISPR loci evolve more rapidly than others, which allowed the strains' phylogenetic relationships to be determined. A comparative genomic analysis showed that E. coli and S. enterica evolve much more slowly than S. thermophilus. The latter's strains that diverged 250 thousand years ago still contained the same spacer complement.[125]Metagenomic analysis of two acid mine drainage biofilms showed that one of the analyzed CRISPRs contained extensive deletions and spacer additions versus the other biofilm, suggesting a higher phage activity/prevalence in one community than the other.[62] In the oral cavity, a temporal study determined that 7-22% of spacers were shared over 17 months within an individual while less than 2% were shared across individuals.[103]
From the same environment a single strain was tracked using PCR primers specific to its CRISPR system. Broad-level results of spacer presence/absence showed significant diversity. However, this CRISPR added 3 spacers over 17 months,[103] suggesting that even in an environment with significant CRISPR diversity some loci evolve slowly.
CRISPRs were analysed from the metagenomes produced for the human microbiome project.[126] Although most were body-site specific, some within a body site are widely shared among individuals. One of these loci originated from streptococcal species and contained ~15,000 spacers, 50% of which were unique. Similar to the targeted studies of the oral cavity, some showed little evolution over time.[126]
CRISPR evolution was studied in chemostats using S. thermophilus to directly examine spacer acquisition rates. In one week, S. thermophilus strains acquired up to three spacers when challenged with a single phage.[127] During the same interval the phage developed single nucleotide polymorphisms that became fixed in the population, suggesting that targeting had prevented phage replication absent these mutations.[127]
Another S. thermophilus experiment showed that phages can infect and replicate in hosts that have only one targeting spacer. Yet another showed that sensitive hosts can exist in environments with high phage titres.[128] The chemostat and observational studies suggest many nuances to CRISPR and phage (co)evolution.
Identification
CRISPRs are widely distributed among bacteria and archaea[70] and show some sequence similarities.[109] Their most notable characteristic is their repeating spacers and direct repeats. This characteristic makes CRISPRs easily identifiable in long sequences of DNA, since the number of repeats decreases the likelihood of a false positive match. Three programs used for CRISPR repeat identification search for regularly interspaced repeats in long sequences: CRT,[129] PILER-CR[130] and CRISPRfinder.[131]Analysis of CRISPRs in metagenomic data is more challenging, as CRISPR loci do not typically assemble, due to their repetitive nature or through strain variation, which confuses assembly algorithms. Where many reference genomes are available, polymerase chain reaction (PCR) can be used to amplify CRISPR arrays and analyse spacer content.[94][103][132][133][134] However, this approach yields information only for specifically targeted CRISPRs and for organisms with sufficient representation in public databases to design reliable polymerase chain reaction (PCR) primers.
The alternative is to extract and reconstruct CRISPR arrays from shotgun metagenomic data. This is computationally more difficult, particularly with second generation sequencing technologies (e.g. 454, Illumina), as the short read lengths prevent more than two or three repeat units appearing in a single read. CRISPR identification in raw reads has been achieved using purely de novo identification[135] or by using direct repeat sequences in partially assembled CRISPR arrays from contigs (overlapping DNA segments that together represent a consensus region of DNA)[126] and direct repeat sequences from published genomes[136] as a hook for identifying direct repeats in individual reads.
Use by phages
Another way for bacteria to defend against phage infection is by having chromosomal islands. A subtype of chromosomal islands called phage-inducible chromosomal island (PICI) is excised from a bacterial chromosome upon phage infection and can inhibit phage replication.[137] The mechanisms that induce PICI excision and how PICI inhibits phage replication are not well understood. One study showed that lytic ICP1 phage, which specifically targets Vibrio cholerae serogroup O1, has acquired a CRISPR/Cas system that targets a V. cholera PICI-like element. The system has 2 CRISPR loci and 9 Cas genes. It seems to be homologous to the 1-F system found in Yersinia pestis. Moreover, like the bacterial CRISPR/Cas system, ICP1 CRISPR/Cas can acquire new sequences, which allows phage and host to co-evolve.[138]Applications
By the end of 2014 some 1000 research papers had been published that mentioned CRISPR.[139][140] The technology had been used to functionally inactivate genes in human cell lines and cells, to study Candida albicans, to modify yeasts used to make biofuels and to genetically modify crop strains.[140] CRISPR can also be used to change mosquitos so they cannot transmit diseases such as malaria.[141]CRISPR-based re-evaluations of claims for gene-disease relationships have led to the discovery of potentially important anomalies.[142]
DNA repair after double-strand break
Predecessors
In the early 2000s, researchers developed zinc finger nucleases (ZFNs), synthetic proteins whose DNA-binding domains enable them to create double-stranded breaks in DNA at specific points. In 2010, synthetic nucleases called transcription activator-like effector nucleases (TALENs) provided an easier way to target a double-stranded break to a specific location on the DNA strand. Both zinc finger nucleases and TALENs require the creation of a custom protein for each targeted DNA sequence, which is a more difficult and time-consuming process than that for guide RNAs. CRISPRs are much easier to design because the process requires making only a short RNA sequence.[143]Whereas RNA interference (RNAi) does not fully suppress gene function, CRISPR, ZFNs and TALENs provide full irreversible gene knockout.[144] CRISPR can also target several DNA sites simultaneously by simply introducing different gRNAs. In addition, CRISPR costs are relatively low.[144][145][146]
Genome engineering
CRISPR/Cas9 genome editing is carried out with a Type II CRISPR system. When utilized for genome editing, this system includes Cas9, crRNA, tracrRNA along with an optional section of DNA repair template that is utilized in either non-homologous end joining (NHEJ) or homology directed repair (HDR).Major components
| Component | Function |
|---|---|
| crRNA | Contains the guide RNA that locates the correct section of host DNA along with a region that binds to tracrRNA (generally in a hairpin loop form) forming an active complex. |
| tracrRNA | Binds to crRNA and forms an active complex. |
| sgRNA | Single guide RNAs are a combined RNA consisting of a tracrRNA and at least one crRNA |
| Cas9 | Protein whose active form is able to modify DNA. Many variants exist with differing functions (i.e. single strand nicking, double strand break, DNA binding) due to Cas9's DNA site recognition function. |
| Repair template | DNA that guides the cellular repair process allowing insertion of a specific DNA sequence |
CRISPR/Cas9 often employs a plasmid to transfect the target cells.[149] The main components of this plasmid are displayed in the image and listed in the table. The crRNA needs to be designed for each application as this is the sequence that Cas9 uses to identify and directly bind to the cell's DNA. The crRNA must bind only where editing is desired. The repair template is designed for each application, as it must overlap with the sequences on either side of the cut and code for the insertion sequence.
Multiple crRNAs and the tracrRNA can be packaged together to form a single-guide RNA (sgRNA).[150] This sgRNA can be joined together with the Cas9 gene and made into a plasmid in order to be transfected into cells.
Overview of the transfection and DNA cleaving by CRISPR Cas9 (crRNA and
tracrRNA are often joined as one strand of RNA when designing a plasmid)[149]
Structure
CRISPR/Cas9 offers a high degree of fidelity and relatively simple construction. It depends on two factors for its specificity: the target sequence and the PAM. The target sequence is 20 bases long as part of each CRISPR locus in the crRNA array.[149] A typical crRNA array has multiple unique target sequences. Cas9 proteins select the correct location on the host's genome by utilizing the sequence to bond with base pairs on the host DNA. The sequence is not part of the Cas9 protein and as a result is customizable and can be independently synthesized.[151][152]The PAM sequence on the host genome is recognized by Cas9. Cas9 cannot be easily modified to recognize a different PAM sequence. However this is not too limiting as it is a short sequence and nonspecific (e.g. the SpCas9 PAM sequence is 5'-NGG-3' and in the human genome occurs roughly every 8 to 12 base pairs).[149]
Once these have been assembled into a plasmid and transfected into cells the Cas9 protein with the help of the crRNA finds the correct sequence in the host cell's DNA and – depending on the Cas9 variant – creates a single or double strand break in the DNA.[153]
Properly spaced single strand breaks in the host DNA can trigger homology directed repair, which is less error prone than the non-homologous end joining that typically follows a double strand break. Providing a DNA repair template allows for the insertion of a specific DNA sequence at an exact location within the genome. The repair template should extend 40 to 90 base pairs beyond the Cas9 induced DNA break.[149] The goal is for the cell's HDR process to utilize the provided repair template and thereby incorporate the new sequence into the genome. Once incorporated, this new sequence is now part of the cell's genetic material and passes into its daughter cells.
Many online tools are available to aid in designing effective sgRNA sequences.[154]
Delivery
Scientists can use viral or non-viral systems for delivery of the Cas9 and sgRNA into target cells. Electroporation of DNA, RNA or ribonucleocomplexes is the most common and cheapest system. This technique was used to edit CXCR4 and PD-1, knocking in new sequences to replace specific genetic "letters" in these proteins. The group was then able to sort the cells, using cell surface markers, to help identify successfully edited cells.[155] Deep sequencing of a target site confirmed that knock-in genome modifications had occurred with up to ∼20% efficiency, which accounted for up to approximately one-third of total editing events.[156] However, hard-to-transfect cells (stem cells, neurons, hematopoietic cells, etc.) require more efficient delivery systems such as those based on lentivirus (LVs), adenovirus (AdV) and adeno-associated virus (AAV).Editing
CRISPRs have been used to cut five[35] to 62 genes at once: pig cells have been engineered to inactivate all 62 Porcine Endogenous Retroviruses in the pig genome, which eliminated transinfection from the pig to human cells in culture.[157] CRISPR's low cost compared to alternatives is widely seen as revolutionary.[11][12]Selective engineered redirection of the CRISPR/Cas system was first demonstrated in 2012 in:[158][159]
- Immunization of industrially important bacteria, including some used in food production and large-scale fermentation
- Cellular or organism RNA-guided genome engineering. Proof of concept studies demonstrated examples both in vitro[13][39][72] and in vivo[51][160][161]
Controlled genome editing
Several variants of CRISPR/Cas9 allow gene activation or genome editing with an external trigger such as light or small molecules.[162][163][164] These include photoactivatable CRISPR systems developed by fusing light-responsive protein partners with an activator domain and a dCas9 for gene activation,[165][166] or fusing similar light responsive domains with two constructs of split-Cas9,[167][168] or by incorporating caged unnatural amino acids into Cas9,[169] or by modifying the guide RNAs with photocleavable complements for genome editing.[170]Methods to control genome editing with small molecules include an allosteric Cas9, with no detectable background editing, that will activate binding and cleavage upon the addition of 4-hydroxytamoxifen (4-HT),[162] 4-HT responsive intein-linked Cas9s[171] or a Cas9 that is 4-HT responsive when fused to four ERT2 domains.[172] Intein-inducible split-Cas9 allows dimerization of Cas9 fragments[173] and Rapamycin-inducible split-Cas9 system developed by fusing two constructs of split Cas9 with FRB and FKBP fragments.[174] Furthermore, other studies have shown to induce transcription of Cas9 with a small molecule, doxycyline.[175][176] Small molecules can also be used to improve Homology Directed Repair (HDR),[177] often by inhibiting the Non-Homologous End Joining (NHEJ) pathway.[178] These systems allow conditional control of CRISPR activity for improved precision, efficiency and spatiotemporal control.
In 2017, researchers successfully used CRISPR-Cas9 as a treatment in a mouse model of human genetic deafness, by genetically editing the DNA in some cells in the ears of live mice.[179]
Knockdown/activation

Using "dead" versions of Cas9 (dCas9) eliminates CRISPR's DNA-cutting ability, while preserving its ability to target desirable sequences. Multiple groups added various regulatory factors to dCas9s, enabling them to turn almost any gene on or off or adjust its level of activity.[180] Like RNAi, CRISPR interference (CRISPRi) turns off genes in a reversible fashion by targeting, but not cutting a site. The targeted site is methylated, epigenetically modifying the gene. This modification inhibits transcription. Conversely, CRISPR-mediated activation (CRISPRa) promotes gene transcription.[181] Cas9 is an effective way of targeting and silencing specific genes at the DNA level.[182] In bacteria, the presence of Cas9 alone is enough to block transcription. For mammalian applications, a section of protein is added. Its guide RNA targets regulatory DNA sequences called promoters that immediately precede the target gene.[35]
Cas9 was used to carry synthetic transcription factors that activated specific human genes. The technique achieved a strong effect by targeting multiple CRISPR constructs to slightly different locations on the gene's promoter.[35]
RNA editing
In 2016 researchers demonstrated that CRISPR from an ordinary mouth bacterium could be used to edit RNA. The researchers searched databases containing hundreds of millions of genetic sequences for those that resembled Crispr genes. They considered the fusobacteria Leptotrichia shahii. It had a group of genes that resembled CRISPR genes, but with important differences. When the researchers equipped other bacteria with these genes, which they called C2c2, they found that the organisms gained a novel defense.[183]Many viruses encode their genetic information in RNA rather than DNA that they repurpose to make new viruses. HIV and poliovirus are such viruses. Bacteria with C2c2 make molecules that can dismember RNA, destroying the virus. Tailoring these genes opened any RNA molecule to editing.[183]
CRISPR-Cas systems can also be employed for editing of micro-RNA and long-noncoding RNA genes in plants.[184]
Disease models
CRISPR simplifies creation of animals for research that mimic disease or show what happens when a gene is knocked down or mutated. CRISPR may be used at the germline level to create animals where the gene is changed everywhere, or it may be targeted at non-germline cells.[185][186][187]CRISPR can be utilized to create human cellular models of disease. For instance, applied to human pluripotent stem cells CRISPR introduced targeted mutations in genes relevant to polycystic kidney disease (PKD) and focal segmental glomerulosclerosis (FSGS).[188] These CRISPR-modified pluripotent stem cells were subsequently grown into human kidney organoids that exhibited disease-specific phenotypes. Kidney organoids from stem cells with PKD mutations formed large, translucent cyst structures from kidney tubules. The cysts were capable of reaching macroscopic dimensions, up to one centimeter in diameter.[189] Kidney organoids with mutations in a gene linked to FSGS developed junctional defects between podocytes, the filtering cells affected in that disease. This was traced to the inability of podocytes ability to form microvilli between adjacent cells.[190] Importantly, these disease phenotypes were absent in control organoids of identical genetic background, but lacking the CRISPR modifications.[188]
A similar approach was taken to model long QT syndrome in cardiomyocytes derived from pluripotent stem cells.[191] These CRISPR-generated cellular models, with isogenic controls, provide a new way to study human disease and test drugs.
Gene drive
Gene drives may provide a powerful tool to restore balance of ecosystems by eliminating invasive species. Concerns regarding efficacy, unintended consequences in the target species as well as non-target species have been raised particularly in the potential for accidental release from laboratories into the wild. Scientists have proposed several safeguards for ensuring the containment of experimental gene drives including molecular, reproductive, and ecological.[192] Many recommend that immunization and reversal drives be developed in tandem with gene drives in order to overwrite their effects if necessary.[193] There remains consensus that long-term effects must be studied more thoroughly particularly in the potential for ecological disruption that cannot be corrected with reversal drives.[194]Biomedicine
CRISPR/Cas-based "RNA-guided nucleases" can be used to target virulence factors, genes encoding antibiotic resistance and other medically relevant sequences of interest. This technology thus represents a novel form of antimicrobial therapy and a strategy by which to manipulate bacterial populations.[195][196] Recent studies suggested a correlation between the interfering of the CRISPR/Cas locus and acquisition of antibiotic resistance[197] This system provides protection of bacteria against invading foreign DNA, such as transposons, bacteriophages and plasmids. This system was shown to be a strong selective pressure for the acquisition of antibiotic resistance and virulence factor in bacterial pathogens.[197] Some of the affected genes are tied to human diseases, including those involved in muscle differentiation, cancer, inflammation and fetal hemoglobin.[35]Research suggests that CRISPR is an effective way to limit replication of multiple herpesviruses. It was able to eradicate viral DNA in the case of Epstein-Barr virus (EBV). Anti-herpesvirus CRISPRs have promising applications such as removing cancer-causing EBV from tumor cells, helping rid donated organs for immunocompromised patients of viral invaders, or preventing cold sore outbreaks and recurrent eye infections by blocking HSV-1 reactivation. As of August 2016, these were awaiting testing.[198] CRISPR is being applied to develop tissue-based treatments for cancer and other diseases.[180][199]
CRISPR may revive the concept of transplanting animal organs into people. Retroviruses present in animal genomes could harm transplant recipients. In 2015 a team eliminated 62 copies of a retrovirus's DNA from the pig genome in a kidney epithelial cell.[180] Researchers recently demonstrated the ability to birth live pig specimens after removing these retroviruses from their genome using CRISPR for the first time.[200]
CRISPR may have applications in tissue engineering and regenerative medicine, such as by creating human blood vessels that lack expression of MHC class II proteins, which often cause transplant rejection.[201]
CRISPR in Cancer
As of 2016 CRISPR had been studied in animal models and cancer cell lines, to learn if it can be used to repair or thwart mutated genes that cause cancer.[202]The first clinical trial involving CRISPR started in 2016. It involved removing immune cells from people with lung cancer, using CRISPR to edit out the gene expressed PD-1, then administrating the altered cells back to the same person. 20 other trials were under way or nearly ready, mostly in China, as of 2017.[203]
In 2016 the United States Food and Drug Administration (FDA) approved a clinical trial in which CRISPR would be used to alter T cells extracted from people with different kinds of cancer and then administer those engineered T cells back to the same people.[204]
Gene function
In 2015, multiple studies attempted to systematically disable each individual human gene, in an attempt to identify which genes were essential to human biology. Between 1,600 and 1,800 genes passed this test—of the 20,000 or so known human genes. Such genes are more strongly activated, and unlikely to carry disabling mutations. They are more likely to have indispensable counterparts in other species. They build proteins that unite to form larger collaborative complexes. The studies also cataloged the essential genes in four cancer-cell lines and identified genes that are expendable in healthy cells, but crucial in specific tumor types and drugs that could target these rogue genes.[205]The specific functions of some 18 percent of the essential genes are unidentified. In one 2015 targeting experiment, disabling individual genes in groups of cells attempted to identify those involved in resistance to a melanoma drug. Each such gene manipulation is itself a separate "drug", potentially opening the entire genome to CRISPR-based regulation.[180]
In 2016-2017, a CRISPR/Cas-based approach to genetically engineering adult rodent brains in vivo was successfully demonstrated.[206][207]
In vitro genetic depletion
Unenriched sequencing libraries often have abundant undesired sequences. Cas9 can specifically deplete the undesired sequences with double strand breakage with up to 99% efficiency and without significant off-target effects as seen with restriction enzymes. Treatment with Cas9 can deplete abundant rRNA while increasing pathogen sensitivity in RNA-seq libraries.[208]In vivo applications
CRISPR/Cas-9 can be used to edit the DNA of organisms in vivo and entire chromosomes can be eliminated from an organism at any point in its development. Chromosomes that have been deleted in vivo are the Y chromosomes and X chromosomes of adult lab mice and human chromosomes 14 and 21, in embryonic stem cell lines and aneuploid mice respectively. This method might be useful for treating genetic aneuploid diseases such as Down Syndrome and intersex disorders.[209]Successful in vivo genome editing using CRISPR/Cas9 has been shown in several model organisms, such as Arabidopsis, Caenorhadbitis elegans, Saccharomyces cerevisiae and Drosophila.[42][43][44][210][49][211] In Drosophila melanogaster, a nuclease-dead Cas9 (dCas9) was confirmed to highly activate transcription in the target genes of Twist and Snail in vivo.[211] For Arabidopsis and C. elegans, experiments focused on CRISPR-Cas induced genome mutations that could be stably inherited by the progeny of these two species.[210][49]
Patents and commercialization
As of December 2014, patent rights to CRISPR were contested. Several companies formed to develop related drugs and research tools.[212] As companies ramp up financing, doubts as to whether CRISPR can be quickly monetized were raised.[213] In February 2017 the US Patent Office ruled on a patent interference case brought by University of California with respect to patents issued to the Broad Institute, and found that the Broad patents, with claims covering the application of CRISPR/cas9 in eukaryotic cells, were distinct from the inventions claimed by University of California.[214][215][216] Shortly after, University of California filed an appeal of this ruling.[217][218]As of November 2013, SAGE Labs (part of Horizon Discovery group) had exclusive rights from one of those companies to produce and sell genetically engineered rats and non-exclusive rights for mouse and rabbit models.[219] By 2015, Thermo Fisher Scientific had licensed intellectual property from ToolGen to develop CRISPR reagent kits.[220]
In March 2017, the European Patent Office (EPO) announced its intention to allow claims to Max-Planck Institute in Berlin, University of California, and University of Vienna,[221][222] and in August 2017, the EPO announced its intention to allow CRISPR claims in a patent application that MilliporeSigma had filed.[221] As of August 2017 the patent situation in Europe was complex, with MilliporeSigma, ToolGen, Vilnius University, and Harvard contending for claims, along with University of California and Broad.[223]
Society and culture
Human germline modification
On Jan 21, 2018, The Wall Street Journal reported that 86 people in China have had their genes edited using CRISPR.[224][225]At least four labs in the US, labs in China and the UK, and a US biotechnology company called Ovascience announced plans for ongoing research to apply CRISPR to human embryos.[226] Scientists, including a CRISPR co-inventor, urged a worldwide moratorium on applying CRISPR to the human germline, especially for clinical use. They said "scientists should avoid even attempting, in lax jurisdictions, germline genome modification for clinical application in humans" until the full implications "are discussed among scientific and governmental organizations".[53][227] These scientists support basic research on CRISPR and do not see CRISPR as developed enough for any clinical use in making heritable changes to humans.[228]
In April 2015, Chinese scientists reported results of an attempt to alter the DNA of non-viable human embryos using CRISPR to correct a mutation that causes beta thalassemia, a lethal heritable disorder.[229][230] The study had previously been rejected by both Nature and Science in part because of ethical concerns.[231] The experiments resulted in changing only some genes, and had off-target effects on other genes. The researchers stated that CRISPR is not ready for clinical application in reproductive medicine.[231] In April 2016 Chinese scientists were reported to have made a second unsuccessful attempt to alter the DNA of non-viable human embryos using CRISPR - this time to alter the CCR5 gene to make the embryo HIV resistant.[232]
In December 2015, an International Summit on Human Gene Editing took place in Washington under the guidance of David Baltimore. Members of national scientific academies of America, Britain and China discussed the ethics of germline modification. They agreed to support basic and clinical research under appropriate legal and ethical guidelines. A specific distinction was made between somatic cells, where the effects of edits are limited to a single individual, versus germline cells, where genome changes could be inherited by future generations. Heritable modifications could have unintended and far-reaching consequences for human evolution, genetically (e.g. gene/environment interactions) and culturally (e.g. Social Darwinism). Altering of gametocytes and embryos to generate inheritable changes in humans was defined to be irresponsible. The group agreed to initiate an international forum to address such concerns and harmonize regulations across countries.[233]
Policy barriers to genetic engineering
Policy regulations for the CRISPR/cas9 system vary around the globe. In February 2016, British scientists were given permission by regulators to genetically modify human embryos by using CRISPR-Cas9 and related techniques. However, researchers were forbidden from implanting the embryos and the embryos were to be destroyed after seven days.[234]The US has an elaborate, interdepartmental regulatory system to evaluate new genetically modified foods and crops. For example, the Agriculture Risk Protection Act of 2000 gives the USDA the authority to oversee the detection, control, eradication, suppression, prevention, or retardation of the spread of plant pests or noxious weeds to protect the agriculture, environment and economy of the US. The act regulates any genetically modified organism that utilizes the genome of a predefined 'plant pest' or any plant not previously categorized.[235] In 2015, Yinong Yang successfully deactivated 16 specific genes in the white button mushroom. Since he had not added any foreign DNA to his organism, the mushroom could not be regulated by the USDA under Section 340.2.[236] Yang's white button mushroom was the first organism genetically modified with the Crispr/cas9 protein system to pass US regulation.[237] In 2016, the USDA sponsored a committee to consider future regulatory policy for upcoming genetic modification techniques. With the help of the US National Academies of Sciences, Engineering and Medicine, special interests groups met on April 15 to contemplate the possible advancements in genetic engineering within the next 5 years and potential policy regulations that would need to come into play.[238] With the emergence of rogue genetic engineers employing the technology, the FDA has begun issuing new regulations.[239]
In China, where social conditions sharply contrast with the west, genetic diseases carry a heavy stigma, individuals with mental and physical disabilities get little government support. Furthermore, there are no religious barriers for the use of genetic modifications to change the genotypes of people.[240] This leaves China with fewer policy barriers to the use of this technology.[241][224]
Recognition
Jennifer Doudna
In 2012 and 2013, CRISPR was a runner-up in Science Magazine's Breakthrough of the Year award. In 2015, it was the winner of that award.[180] CRISPR was named as one of MIT Technology Review's 10 breakthrough technologies in 2014 and 2016.[242][243] In 2016, Jennifer Doudna, Emmanuelle Charpentier, along with Rudolph Barrangou, Philippe Horvath, and Feng Zhang won the Gairdner International award. In 2017, Jennifer Doudna and Emmanuelle Charpentier were awarded the Japan Prize for their revolutionary invention of CRISPR-Cas9 in Tokyo, Japan.