Transcriptomics technologies are the techniques used to study an organism’s transcriptome, the sum of all of its RNA transcripts. The information content of an organism is recorded in the DNA of its genome and expressed through transcription. Here, mRNA serves as a transient intermediary molecule in the information network, whilst non-coding RNAs perform additional diverse functions. A transcriptome captures a snapshot in time of the total transcripts present in a cell.
Transcriptomics technologies provide a broad account of which cellular
processes are active and which are dormant.
A major challenge in molecular biology lies in understanding how the
same genome can give rise to different cell types and how gene
expression is regulated.
The first attempts to study whole transcriptomes began in the early 1990s. Subsequent technological advances since the late 1990s have repeatedly transformed the field, and made transcriptomics a widespread discipline in biological sciences. There are two key contemporary techniques in the field: microarrays, which quantify a set of predetermined sequences, and RNA-Seq, which uses high-throughput sequencing to record all transcripts. As the technology improved, the volume of data produced by each transcriptome experiment increased. As a result, data analysis methods have steadily been adapted to more accurately and efficiently analyse increasingly large volumes of data. Transcriptome databases have grown and increased in utility as more transcriptomes are collected and shared by researchers. It would be almost impossible to interpret the information contained in a transcriptome without the context of previous experiments.
Measuring the expression of an organism’s genes in different tissues or conditions, or at different times, gives information on how genes are regulated and reveal details of an organism’s biology. It can also be used to infer the functions of previously unannotated genes. Transcriptome analysis has enabled the study of how gene expression changes in different organisms and has been instrumental in the understanding of human disease. An analysis of gene expression in its entirety allows detection of broad coordinated trends which cannot be discerned by more targeted assays.
The first attempts to study whole transcriptomes began in the early 1990s. Subsequent technological advances since the late 1990s have repeatedly transformed the field, and made transcriptomics a widespread discipline in biological sciences. There are two key contemporary techniques in the field: microarrays, which quantify a set of predetermined sequences, and RNA-Seq, which uses high-throughput sequencing to record all transcripts. As the technology improved, the volume of data produced by each transcriptome experiment increased. As a result, data analysis methods have steadily been adapted to more accurately and efficiently analyse increasingly large volumes of data. Transcriptome databases have grown and increased in utility as more transcriptomes are collected and shared by researchers. It would be almost impossible to interpret the information contained in a transcriptome without the context of previous experiments.
Measuring the expression of an organism’s genes in different tissues or conditions, or at different times, gives information on how genes are regulated and reveal details of an organism’s biology. It can also be used to infer the functions of previously unannotated genes. Transcriptome analysis has enabled the study of how gene expression changes in different organisms and has been instrumental in the understanding of human disease. An analysis of gene expression in its entirety allows detection of broad coordinated trends which cannot be discerned by more targeted assays.
History
Transcriptomics method use over time.
Published papers referring to RNA-Seq (black), RNA microarray (red),
expressed sequence tag (blue) and serial/cap analysis of gene expression
(yellow) since 1990.
Transcriptomics has been characterised by the development of new
techniques which have redefined what is possible every decade or so and
rendered previous technologies obsolete. The first attempt at capturing a
partial human transcriptome was published in 1991 and reported 609 mRNA sequences from the human brain. In 2008, two human transcriptomes, composed of millions of transcript-derived sequences covering 16,000 genes, were published, and by 2015 transcriptomes had been published for hundreds of individuals. Transcriptomes of different disease states, tissues, or even single cells are now routinely generated.
This explosion in transcriptomics has been driven by the rapid
development of new technologies with improved sensitivity and economy.
Before transcriptomics
Studies of individual transcripts were being performed several decades before any transcriptomics approaches were available. Libraries of silkmoth mRNA transcripts were collected and converted to complementary DNA (cDNA) for storage using reverse transcriptase in the late 1970s. In the 1980s, low-throughput sequencing using the Sanger method was used to sequence random transcripts, producing expressed sequence tags (ESTs). The Sanger method of sequencing was predominant until the advent of high-throughput methods such as sequencing by synthesis (Solexa/Illumina). ESTs came to prominence during the 1990s as an efficient method to determine the gene content of an organism without sequencing the entire genome. Amounts of individual transcripts were quantified using Northern blotting, nylon membrane arrays, and later reverse transcriptase quantitative PCR (RT-qPCR) methods, but these methods are laborious and can only capture a tiny subsection of a transcriptome.
Consequently, the manner in which a transcriptome as a whole is
expressed and regulated remained unknown until higher-throughput
techniques were developed.
Early attempts
The word "transcriptome" was first used in the 1990s. In 1995, one of the earliest sequencing-based transcriptomic methods was developed, serial analysis of gene expression (SAGE), which worked by Sanger sequencing of concatenated random transcript fragments.
Transcripts were quantified by matching the fragments to known genes. A
variant of SAGE using high-throughput sequencing techniques, called
digital gene expression analysis, was also briefly used.
However, these methods were largely overtaken by high throughput
sequencing of entire transcripts, which provided additional information
on transcript structure such as splice variants.
Development of contemporary techniques
|
|
RNA-Seq | Microarray |
|---|---|---|
| Throughput | 1 day to 1 week per experiment | 1–2 days per experiment |
| Input RNA amount | Low ~ 1 ng total RNA | High ~ 1 μg mRNA |
| Labour intensity | High (sample preparation and data analysis) | Low |
| Prior knowledge | None required, although a reference genome/transcriptome sequence is useful | Reference genome/transcriptome is required for design of probes |
| Quantitation accuracy | ~90% (limited by sequence coverage) | >90% (limited by fluorescence detection accuracy) |
| Sequence resolution | RNA-Seq can detect SNPs and splice variants (limited by sequencing accuracy of ~99%) | Specialised arrays can detect mRNA splice variants (limited by probe design and cross-hybridisation) |
| Sensitivity | 1 transcript per million (approximate, limited by sequence coverage) | 1 transcript per thousand (approximate, limited by fluorescence detection) |
| Dynamic range | 100,000:1 (limited by sequence coverage) | 1,000:1 (limited by fluorescence saturation) |
| Technical reproducibility | >99% | >99% |
The dominant contemporary techniques, microarrays and RNA-Seq, were developed in the mid-1990s and 2000s. Microarrays that measure the abundances of a defined set of transcripts via their hybridisation to an array of complementary probes were first published in 1995.
Microarray technology allowed the assay of thousands of transcripts
simultaneously and at a greatly reduced cost per gene and labour saving. Both spotted oligonucleotide arrays and Affymetrix high-density arrays were the method of choice for transcriptional profiling until the late 2000s. Over this period, a range of microarrays were produced to cover known genes in model
or economically important organisms. Advances in design and manufacture
of arrays improved the specificity of probes and allowed more genes to
be tested on a single array. Advances in fluorescence detection increased the sensitivity and measurement accuracy for low abundance transcripts.
RNA-Seq is accomplished by reverse transcribing RNA in vitro and sequencing the resulting cDNAs.
Transcript abundance is derived from the number of counts from each
transcript. The technique has therefore been heavily influenced by the
development of high-throughput sequencing technologies. Massively parallel signature sequencing (MPSS) was an early example based on generating 16–20 bp sequences via a complex series of hybridisations, and was used in 2004 to validate the expression of ten thousand genes in Arabidopsis thaliana. The earliest RNA-Seq work was published in 2006 with one hundred thousand transcripts sequenced using 454 technology.
This was sufficient coverage to quantify relative transcript abundance.
RNA-Seq began to increase in popularity after 2008 when new Solexa/Illumina technologies allowed one billion transcript sequences to be recorded. This yield now allows for the quantification and comparison of human transcriptomes.
Data gathering
Generating data on RNA transcripts can be achieved via either of two main principles: sequencing of individual transcripts (ESTs, or RNA-Seq) or hybridisation of transcripts to an ordered array of nucleotide probes (microarrays).
Isolation of RNA
All
transcriptomic methods require RNA to first be isolated from the
experimental organism before transcripts can be recorded. Although
biological systems are incredibly diverse, RNA extraction techniques are broadly similar and involve mechanical disruption of cells or tissues, disruption of RNase with chaotropic salts, disruption of macromolecules and nucleotide complexes, separation of RNA from undesired biomolecules including DNA, and concentration of the RNA via precipitation from solution or elution from a solid matrix. Isolated RNA may additionally be treated with DNase to digest any traces of DNA. It is necessary to enrich messenger RNA as total RNA extracts are typically 98% ribosomal RNA. Enrichment for transcripts can be performed by poly-A affinity methods or by depletion of ribosomal RNA using sequence-specific probes. Degraded RNA may affect downstream results; for example, mRNA enrichment from degraded samples will result in the depletion of 5’ mRNA ends and an uneven signal across the length of a transcript. Snap-freezing of tissue prior to RNA isolation is typical, and care is taken to reduce exposure to RNase enzymes once isolation is complete.
Expressed sequence tags
An expressed sequence tag (EST) is a short nucleotide sequence generated from a single RNA transcript. RNA is first copied as complementary DNA (cDNA) by a reverse transcriptase enzyme before the resultant cDNA is sequenced.
Because ESTs can be collected without prior knowledge of the organism
from which they come, they can be made from mixtures of organisms or
environmental samples. Although higher-throughput methods are now used, EST libraries commonly provided sequence information for early microarray designs; for example, a barley microarray was designed from 350,000 previously sequenced ESTs.
Serial and cap analysis of gene expression (SAGE/CAGE)
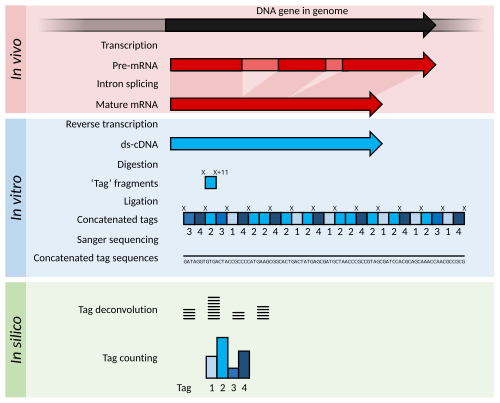
Summary of SAGE. Within the organisms, genes are transcribed and spliced (in eukaryotes) to produce mature mRNA transcripts (red). The mRNA is extracted from the organism, and reverse transcriptase is used to copy the mRNA into stable double-stranded–cDNA (ds-cDNA; blue). In SAGE, the ds-cDNA is digested by restriction enzymes
(at location ‘X’ and ‘X’+11) to produce 11-nucleotide "tag" fragments.
These tags are concatenated and sequenced using long-read Sanger sequencing (different shades of blue indicate tags from different genes). The sequences are deconvoluted to find the frequency of each tag. The tag frequency can be used to report on transcription of the gene that the tag came from.
Serial analysis of gene expression
(SAGE) was a development of EST methodology to increase the throughput
of the tags generated and allow some quantitation of transcript
abundance. cDNA is generated from the RNA but is then digested into 11 bp "tag" fragments using restriction enzymes that cut DNA at a specific sequence, and 11 base pairs along from that sequence. These cDNA tags are then joined head-to-tail into long strands (>500 bp) and sequenced using low-throughput, but long read-length methods such as Sanger sequencing. The sequences are then divided back into their original 11 bp tags using computer software in a process called deconvolution. If a reference genome
is available, these tags may be matched to their corresponding gene in
the genome. If a reference genome is unavailable, the tags can be
directly used as diagnostic markers if found to be differentially expressed in a disease state.
The cap analysis gene expression (CAGE) method is a variant of SAGE that sequences tags from the 5’ end of an mRNA transcript only. Therefore, the transcriptional start site of genes can be identified when the tags are aligned to a reference genome. Identifying gene start sites is of use for promoter analysis and for the cloning of full-length cDNAs.
SAGE and CAGE methods produce information on more genes than was
possible when sequencing single ESTs, but sample preparation and data
analysis are typically more labour-intensive.
Microarrays
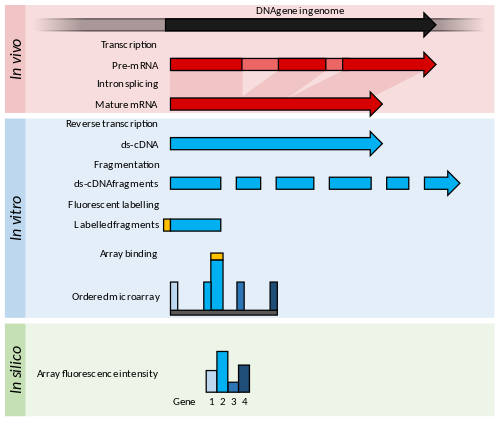
Summary of DNA Microarrays.
Within the organisms, genes are transcribed and spliced (in eukaryotes)
to produce mature mRNA transcripts (red). The mRNA is extracted from
the organism and reverse transcriptase is used to copy the mRNA into
stable ds-cDNA (blue). In microarrays, the ds-cDNA is fragmented and
fluorescently labelled (orange). The labelled fragments bind to an
ordered array of complementary oligonucleotides, and measurement of fluorescent intensity
across the array indicates the abundance of a predetermined set of
sequences. These sequences are typically specifically chosen to report
on genes of interest within the organism's genome.
Principles and advances
Microarrays consist of short nucleotide oligomers, known as "probes", which are typically arrayed in a grid on a glass slide. Transcript abundance is determined by hybridisation of fluorescently labelled transcripts to these probes. The fluorescence intensity at each probe location on the array indicates the transcript abundance for that probe sequence.
Microarrays require some genomic knowledge from the organism of interest, for example, in the form of an annotated genome sequence, or a library of ESTs that can be used to generate the probes for the array.
Methods
Microarrays
for transcriptomics typically fall into one of two broad categories:
low-density spotted arrays or high-density short probe arrays.
Transcript abundance is inferred from the intensity of fluorescence
derived from fluorophore-tagged transcripts that bind to the array.
Spotted low-density arrays typically feature picolitre drops of a range of purified cDNAs arrayed on the surface of a glass slide. These probes are longer than those of high-density arrays and cannot identify alternative splicing events. Spotted arrays use two different fluorophores to label the test and control samples, and the ratio of fluorescence is used to calculate a relative measure of abundance. High-density arrays use a single fluorescent label, and each sample is hybridised and detected individually. High-density arrays were popularised by the Affymetrix GeneChip array, where each transcript is quantified by several short 25-mer probes that together assay one gene.
NimbleGen arrays were a high-density array produced by a maskless-photochemistry
method, which permitted flexible manufacture of arrays in small or
large numbers. These arrays had 100,000s of 45 to 85-mer probes and were
hybridised with a one-colour labelled sample for expression analysis. Some designs incorporated up to 12 independent arrays per slide.
RNA-Seq
Summary of RNA-Seq.
Within the organisms, genes are transcribed and spliced (in eukaryotes)
to produce mature mRNA transcripts (red). The mRNA is extracted from
the organism, fragmented and copied into stable ds-cDNA (blue). The
ds-cDNA is sequenced using high-throughput, short-read sequencing methods. These sequences can then be aligned
to a reference genome sequence to reconstruct which genome regions were
being transcribed. This data can be used to annotate where expressed
genes are, their relative expression levels, and any alternative splice
variants.
Principles and advances
RNA-Seq refers to the combination of a high-throughput sequencing methodology with computational methods to capture and quantify transcripts present in an RNA extract.
The nucleotide sequences generated are typically around 100 bp in
length, but can range from 30 bp to over 10,000 bp depending on the
sequencing method used. RNA-Seq leverages deep sampling
of the transcriptome with many short fragments from a transcriptome to
allow computational reconstruction of the original RNA transcript by aligning reads to a reference genome or to each other (de novo assembly). Both low-abundance and high-abundance RNAs can be quantified in an RNA-Seq experiment (dynamic range of 5 orders of magnitude)—a
key advantage over microarray transcriptomes. In addition, input RNA
amounts are much lower for RNA-Seq (nanogram quantity) compared to
microarrays (microgram quantity), which allows finer examination of
cellular structures down to the single-cell level when combined with
linear amplification of cDNA.
Theoretically, there is no upper limit of quantification in RNA-Seq,
and background noise is very low for 100 bp reads in non-repetitive
regions.
RNA-Seq may be used to identify genes within a genome,
or identify which genes are active at a particular point in time, and
read counts can be used to accurately model the relative gene expression
level. RNA-Seq methodology has constantly improved, primarily through
the development of DNA sequencing technologies to increase throughput,
accuracy, and read length. Since the first descriptions in 2006 and 2008, RNA-Seq has been rapidly adopted and overtook microarrays as the dominant transcriptomics technique in 2015.
The quest for transcriptome data at the level of individual cells
has driven advances in RNA-Seq library preparation methods, resulting
in dramatic advances in sensitivity. Single-cell transcriptomes are now well described and have even been extended to in situ RNA-Seq where transcriptomes of individual cells are directly interrogated in fixed tissues.
Methods
RNA-Seq was established in concert with the rapid development of a range of high-throughput DNA sequencing technologies.
However, before the extracted RNA transcripts are sequenced, several
key processing steps are performed. Methods differ in the use of
transcript enrichment, fragmentation, amplification, single or
paired-end sequencing, and whether to preserve strand information.
The sensitivity of an RNA-Seq experiment can be increased by
enriching classes of RNA that are of interest and depleting known
abundant RNAs. The mRNA molecules can be separated using
oligonucleotides probes which bind their poly-A tails. Alternatively, ribo-depletion can be used to specifically remove abundant but uninformative ribosomal RNAs (rRNAs) by hybridisation to probes tailored to the taxon's
specific rRNA sequences (e.g. mammal rRNA, plant rRNA). However,
ribo-depletion can also introduce some bias via non-specific depletion
of off-target transcripts. Small RNAs, such as micro RNAs, can be purified based on their size by gel electrophoresis and extraction.
Since mRNAs are longer than the read-lengths of typical
high-throughput sequencing methods, transcripts are usually fragmented
prior to sequencing. The fragmentation method is a key aspect of sequencing library construction. Fragmentation may be achieved by chemical hydrolysis, nebulisation, sonication, or reverse transcription with chain-terminating nucleotides. Alternatively, fragmentation and cDNA tagging may be done simultaneously by using transposase enzymes.
During preparation for sequencing, cDNA copies of transcripts may be amplified by PCR to enrich for fragments that contain the expected 5’ and 3’ adapter sequences. Amplification is also used to allow sequencing of very low input amounts of RNA, down to as little as 50 pg in extreme applications. Spike-in controls of known RNAs can be used for quality control assessment to check library preparation and sequencing, in terms of GC-content, fragment length, as well as the bias due to fragment position within a transcript. Unique molecular identifiers
(UMIs) are short random sequences that are used to individually tag
sequence fragments during library preparation so that every tagged
fragment is unique.
UMIs provide an absolute scale for quantification, the opportunity to
correct for subsequent amplification bias introduced during library
construction, and accurately estimate the initial sample size. UMIs are
particularly well-suited to single-cell RNA-Seq transcriptomics, where
the amount of input RNA is restricted and extended amplification of the
sample is required.
Once the transcript molecules have been prepared they can be
sequenced in just one direction (single-end) or both directions
(paired-end). A single-end sequence is usually quicker to produce,
cheaper than paired-end sequencing and sufficient for quantification of
gene expression levels. Paired-end sequencing produces more robust
alignments/assemblies, which is beneficial for gene annotation and
transcript isoform discovery. Strand-specific RNA-Seq methods preserve the strand information of a sequenced transcript.
Without strand information, reads can be aligned to a gene locus but do
not inform in which direction the gene is transcribed. Stranded-RNA-Seq
is useful for deciphering transcription for genes that overlap in different directions and to make more robust gene predictions in non-model organisms.
| Platform | Commercial release | Typical read length | Maximum throughput per run | Single read accuracy | RNA-Seq runs deposited in the NCBI SRA (Oct 2016) |
|---|---|---|---|---|---|
| 454 Life Sciences | 2005 | 700 bp | 0.7 Gbp | 99.9% | 3548 |
| Illumina | 2006 | 50–300 bp | 900 Gbp | 99.9% | 362903 |
| SOLiD | 2008 | 50 bp | 320 Gbp | 99.9% | 7032 |
| Ion Torrent | 2010 | 400 bp | 30 Gbp | 98% | 1953 |
| PacBio | 2011 | 10,000 bp | 2 Gbp | 87% | 160 |
Legend: NCBI SRA – National center for biotechnology information sequence read archive.
Currently RNA-Seq relies on copying RNA molecules into cDNA
molecules prior to sequencing; therefore, the subsequent platforms are
the same for transcriptomic and genomic data. Consequently the
development of DNA sequencing technologies has been a defining feature
of RNA-Seq. Direct sequencing of RNA using nanopore sequencing represents a current state-of-the-art RNA-Seq technique. Nanopore sequencing of RNA can detect modified bases that would be otherwise masked when sequencing cDNA and also eliminates amplification steps that can otherwise introduce bias.
The sensitivity and accuracy of an RNA-Seq experiment are dependent on the number of reads obtained from each sample.
A large number of reads are needed to ensure sufficient coverage of the
transcriptome, enabling detection of low abundance transcripts.
Experimental design is further complicated by sequencing technologies
with a limited output range, the variable efficiency of sequence
creation, and variable sequence quality. Added to those considerations
is that every species has a different number of genes
and therefore requires a tailored sequence yield for an effective
transcriptome. Early studies determined suitable thresholds empirically,
but as the technology matured suitable coverage was predicted
computationally by transcriptome saturation. Somewhat
counter-intuitively, the most effective way to improve detection of
differential expression in low expression genes is to add more biological replicates rather than adding more reads. The current benchmarks recommended by the Encyclopedia of DNA Elements
(ENCODE) Project are for 70-fold exome coverage for standard RNA-Seq
and up to 500-fold exome coverage to detect rare transcripts and
isoforms.
Data analysis
Transcriptomics
methods are highly parallel and require significant computation to
produce meaningful data for both microarray and RNA-Seq experiments. Microarray data is recorded as high-resolution images, requiring feature detection and spectral analysis.
Microarray raw image files are each about 750 MB in size, while the
processed intensities are around 60 MB in size. Multiple short probes
matching a single transcript can reveal details about the intron-exon
structure, requiring statistical models to determine the authenticity
of the resulting signal. RNA-Seq studies produce billions of short DNA
sequences, which must be aligned to reference genomes composed of millions to billions of base pairs. De novo assembly of reads within a dataset requires the construction of highly complex sequence graphs. RNA-Seq operations are highly repetitious and benefit from parallelised computation
but modern algorithms mean consumer computing hardware is sufficient
for simple transcriptomics experiments that do not require de novo assembly of reads. A human transcriptome could be accurately captured using RNA-Seq with 30 million 100 bp sequences per sample. This example would require approximately 1.8 gigabytes of disk space per sample when stored in a compressed fastq format.
Processed count data for each gene would be much smaller, equivalent to
processed microarray intensities. Sequence data may be stored in public
repositories, such as the Sequence Read Archive (SRA). RNA-Seq datasets can be uploaded via the Gene Expression Omnibus.
Image processing
Microarray and sequencing flow cell.
Microarrays and RNA-seq rely on image analysis in different ways. In a
microarray chip, each spot on a chip is a defined oligonucleotide probe,
and fluorescence intensity directly detects the abundance of a specific
sequence (Affymetrix). In a high-throughput sequencing flow cell, spots
are sequenced one nucleotide at a time, with the colour at each round
indicating the next nucleotide in the sequence (Illumina Hiseq). Other
variations of these techniques use more or fewer colour channels.
Microarray image processing must correctly identify the regular grid of features within an image and independently quantify the fluorescence intensity for each feature. Image artefacts
must be additionally identified and removed from the overall analysis.
Fluorescence intensities directly indicate the abundance of each
sequence, since the sequence of each probe on the array is already
known.
The first steps of RNA-seq also include similar image processing;
however, conversion of images to sequence data is typically handled
automatically by the instrument software. The Illumina
sequencing-by-synthesis method results in an array of clusters
distributed over the surface of a flow cell.
The flow cell is imaged up to four times during each sequencing cycle,
with tens to hundreds of cycles in total. Flow cell clusters are
analogous to microarray spots and must be correctly identified during
the early stages of the sequencing process. In Roche’s pyrosequencing
method, the intensity of emitted light determines the number of
consecutive nucleotides in a homopolymer repeat. There are many variants
on these methods, each with a different error profile for the resulting
data.
RNA-Seq data analysis
RNA-Seq
experiments generate a large volume of raw sequence reads which have to
be processed to yield useful information. Data analysis usually
requires a combination of bioinformatics software tools
that vary according to the experimental design and goals. The process
can be broken down into four stages: quality control, alignment,
quantification, and differential expression. Most popular RNA-Seq programs are run from a command-line interface, either in a Unix environment or within the R/Bioconductor statistical environment.
Quality control
Sequence
reads are not perfect, so the accuracy of each base in the sequence
needs to be estimated for downstream analyses. Raw data is examined to
ensure: quality scores for base calls are high, the GC content matches
the expected distribution, short sequence motifs (k-mers) are not over-represented, and the read duplication rate is acceptably low. Several software options exist for sequence quality analysis, including FastQC and FaQCs. Abnormalities may be removed (trimming) or tagged for special treatment during later processes.
Alignment
In order to link sequence read abundance to the expression of a particular gene, transcript sequences are aligned to a reference genome or de novo aligned to one another if no reference is available. The key challenges for alignment software
include sufficient speed to permit billions of short sequences to be
aligned in a meaningful timeframe, flexibility to recognise and deal
with intron splicing of eukaryotic mRNA, and correct assignment of reads
that map to multiple locations. Software advances have greatly
addressed these issues, and increases in sequencing read length reduce
the chance of ambiguous read alignments. A list of currently available
high-throughput sequence aligners is maintained by the EBI.
Alignment of primary transcript mRNA sequences derived from eukaryotes to a reference genome requires specialised handling of intron sequences, which are absent from mature mRNA. Short read aligners perform an additional round of alignments specifically designed to identify splice junctions,
informed by canonical splice site sequences and known intron splice
site information. Identification of intron splice junctions prevents
reads from being misaligned across splice junctions or erroneously
discarded, allowing more reads to be aligned to the reference genome and
improving the accuracy of gene expression estimates. Since gene regulation may occur at the mRNA isoform
level, splice-aware alignments also permit detection of isoform
abundance changes that would otherwise be lost in a bulked analysis.
De novo assembly can be used to align reads to one another
to construct full-length transcript sequences without use of a
reference genome. Challenges particular to de novo
assembly include larger computational requirements compared to a
reference-based transcriptome, additional validation of gene variants or
fragments, and additional annotation of assembled transcripts. The
first metrics used to describe transcriptome assemblies, such as N50, have been shown to be misleading and improved evaluation methods are now available. Annotation-based metrics are better assessments of assembly completeness, such as contig reciprocal best hit count. Once assembled de novo, the assembly can be used as a reference for subsequent sequence alignment methods and quantitative gene expression analysis.
| Software | Released | Last updated | Computational efficiency | Strengths and weaknesses |
|---|---|---|---|---|
| Velvet-Oases | 2008 | 2011 | Low, single-threaded, high RAM requirement | The original short read assembler. It is now largely superseded. |
| SOAPdenovo-trans | 2011 | 2014 | Moderate, multi-thread, medium RAM requirement | An early example of a short read assembler. It has been updated for transcriptome assembly. |
| Trans-ABySS | 2010 | 2016 | Moderate, multi-thread, medium RAM requirement | Suited to short reads, can handle complex transcriptomes, and an MPI-parallel version is available for computing clusters. |
| Trinity | 2011 | 2017 | Moderate, multi-thread, medium RAM requirement | Suited to short reads. It can handle complex transcriptomes but is memory intensive. |
| miraEST | 1999 | 2016 | Moderate, multi-thread, medium RAM requirement | Can process repetitive sequences, combine different sequencing formats, and a wide range of sequence platforms are accepted. |
| Newbler | 2004 | 2012 | Low, single-thread, high RAM requirement | Specialised to accommodate the homo-polymer sequencing errors typical of Roche 454 sequencers. |
| CLC genomics workbench | 2008 | 2014 | High, multi-thread, low RAM requirement | Has a graphical user interface, can combine diverse sequencing technologies, has no transcriptome-specific features, and a licence must be purchased before use. |
| SPAdes | 2012 | 2017 | High, multi-thread, low RAM requirement | Used for transcriptomics experiments on single cells. |
| RSEM | 2011 | 2017 | High, multi-thread, low RAM requirement | Can estimate frequency of alternatively spliced transcripts. User friendly. |
| StringTie | 2015 | 2019 | High, multi-thread, low RAM requirement | Can use a combination of reference-guided and de novo assembly methods to identify transcripts. |
Legend: RAM – random access memory; MPI – message passing interface; EST – expressed sequence tag.
Quantification
Heatmap identification of gene co-expression patterns across different samples.
Each column contains the measurements for gene expression change for a
single sample. Relative gene expression is indicated by colour:
high-expression (red), median-expression (white) and low-expression
(blue). Genes and samples with similar expression profiles can be
automatically grouped (left and top trees). Samples may be different
individuals, tissues, environments or health conditions. In this
example, expression of gene set 1 is high and expression of gene set 2
is low in samples 1, 2, and 3.
Quantification of sequence alignments may be performed at the gene, exon, or transcript level. Typical outputs include a table of read counts for each feature supplied to the software; for example, for genes in a general feature format file. Gene and exon read counts may be calculated quite easily using HTSeq, for example.
Quantitation at the transcript level is more complicated and requires
probabilistic methods to estimate transcript isoform abundance from
short read information; for example, using cufflinks software.
Reads that align equally well to multiple locations must be identified
and either removed, aligned to one of the possible locations, or aligned
to the most probable location.
Some quantification methods can circumvent the need for an exact
alignment of a read to a reference sequence altogether. The kallisto
software method combines pseudoalignment and quantification into a
single step that runs 2 orders of magnitude faster than contemporary
methods such as those used by tophat/cufflinks software, with less
computational burden.
Differential expression
Once quantitative counts of each transcript are available, differential gene expression is measured by normalising, modelling, and statistically analysing the data. Most tools will read a table of genes and read counts as their input, but some programs, such as cuffdiff, will accept binary alignment map
format read alignments as input. The final outputs of these analyses
are gene lists with associated pair-wise tests for differential
expression between treatments and the probability estimates of those
differences.
| Software | Environment | Specialisation |
|---|---|---|
| Cuffdiff2 | Unix-based | Transcript analysis that tracks alternative splicing of mRNA |
| EdgeR | R/Bioconductor | Any count-based genomic data |
| DEseq2 | R/Bioconductor | Flexible data types, low replication |
| Limma/Voom | R/Bioconductor | Microarray or RNA-Seq data, flexible experiment design |
| Ballgown | R/Bioconductor | Efficient and sensitive transcript discovery, flexible. |
Legend: mRNA - messenger RNA.
Validation
Transcriptomic analyses may be validated using an independent technique, for example, quantitative PCR (qPCR), which is recognisable and statistically assessable. Gene expression is measured against defined standards both for the gene of interest and control
genes. The measurement by qPCR is similar to that obtained by RNA-Seq
wherein a value can be calculated for the concentration of a target
region in a given sample. qPCR is, however, restricted to amplicons smaller than 300 bp, usually toward the 3’ end of the coding region, avoiding the 3’UTR. If validation of transcript isoforms is required, an inspection of RNA-Seq read alignments should indicate where qPCR primers
might be placed for maximum discrimination. The measurement of multiple
control genes along with the genes of interest produces a stable
reference within a biological context. qPCR validation of RNA-Seq data has generally shown that different RNA-Seq methods are highly correlated.
Functional validation of key genes is an important consideration
for post transcriptome planning. Observed gene expression patterns may
be functionally linked to a phenotype by an independent knock-down/rescue study in the organism of interest.
Applications
Diagnostics and disease profiling
Transcriptomic strategies have seen broad application across diverse areas of biomedical research, including disease diagnosis and profiling. RNA-Seq approaches have allowed for the large-scale identification of transcriptional start sites, uncovered alternative promoter usage, and novel splicing alterations. These regulatory elements are important in human disease and, therefore, defining such variants is crucial to the interpretation of disease-association studies. RNA-Seq can also identify disease-associated single nucleotide polymorphisms (SNPs), allele-specific expression, and gene fusions, which contributes to the understanding of disease causal variants.
Retrotransposons are transposable elements which proliferate within eukaryotic genomes through a process involving reverse transcription.
RNA-Seq can provide information about the transcription of endogenous
retrotransposons that may influence the transcription of neighboring
genes by various epigenetic mechanisms that lead to disease. Similarly, the potential for using RNA-Seq to understand immune-related disease is expanding rapidly due to the ability to dissect immune cell populations and to sequence T cell and B cell receptor repertoires from patients.
Human and pathogen transcriptomes
RNA-Seq of human pathogens has become an established method for quantifying gene expression changes, identifying novel virulence factors, predicting antibiotic resistance, and unveiling host-pathogen immune interactions. A primary aim of this technology is to develop optimised infection control measures and targeted individualised treatment.
Transcriptomic analysis has predominantly focused on either the
host or the pathogen. Dual RNA-Seq has been applied to simultaneously
profile RNA expression in both the pathogen and host throughout the
infection process. This technique enables the study of the dynamic
response and interspecies gene regulatory networks
in both interaction partners from initial contact through to invasion
and the final persistence of the pathogen or clearance by the host
immune system.
Responses to environment
Transcriptomics allows identification of genes and pathways that respond to and counteract biotic and abiotic environmental stresses.
The non-targeted nature of transcriptomics allows the identification of
novel transcriptional networks in complex systems. For example,
comparative analysis of a range of chickpea lines at different developmental stages identified distinct transcriptional profiles associated with drought and salinity stresses, including identifying the role of transcript isoforms of AP2-EREBP. Investigation of gene expression during biofilm formation by the fungal pathogen Candida albicans revealed a co-regulated set of genes critical for biofilm establishment and maintenance.
Transcriptomic profiling also provides crucial information on mechanisms of drug resistance. Analysis of over 1000 isolates of Plasmodium falciparum, a virulent parasite responsible for malaria in humans, identified that upregulation of the unfolded protein response and slower progression through the early stages of the asexual intraerythrocytic developmental cycle were associated with artemisinin resistance in isolates from Southeast Asia.
Gene function annotation
All transcriptomic techniques have been particularly useful in identifying the functions of genes and identifying those responsible for particular phenotypes. Transcriptomics of Arabidopsis ecotypes that hyperaccumulate metals correlated genes involved in metal uptake, tolerance, and homeostasis with the phenotype.
Integration of RNA-Seq datasets across different tissues has been used
to improve annotation of gene functions in commercially important
organisms (e.g. cucumber) or threatened species (e.g. koala).
Assembly of RNA-Seq reads is not dependent on a reference genome
and so is ideal for gene expression studies of non-model organisms with
non-existing or poorly developed genomic resources. For example, a
database of SNPs used in Douglas fir breeding programs was created by de novo transcriptome analysis in the absence of a sequenced genome.
Similarly, genes that function in the development of cardiac, muscle,
and nervous tissue in lobsters were identified by comparing the
transcriptomes of the various tissue types without use of a genome
sequence. RNA-Seq can also be used to identify previously unknown protein coding regions in existing sequenced genomes.
Non-coding RNA
Transcriptomics
is most commonly applied to the mRNA content of the cell. However, the
same techniques are equally applicable to non-coding RNAs (ncRNAs) that
are not translated into a protein, but instead have direct functions
(e.g. roles in protein translation, DNA replication, RNA splicing, and transcriptional regulation). Many of these ncRNAs affect disease states, including cancer, cardiovascular, and neurological diseases.
Transcriptome databases
Transcriptomics
studies generate large amounts of data that have potential applications
far beyond the original aims of an experiment. As such, raw or
processed data may be deposited in public databases
to ensure their utility for the broader scientific community. For
example, as of 2018, the Gene Expression Omnibus contained millions of
experiments.
| Name | Host | Data | Description |
|---|---|---|---|
| Gene Expression Omnibus | NCBI | Microarray RNA-Seq | First transcriptomics database to accept data from any source. Introduced MIAME and MINSEQE community standards that define necessary experiment metadata to ensure effective interpretation and repeatability. |
| ArrayExpress | ENA | Microarray | Imports datasets from the Gene Expression Omnibus and accepts direct submissions. Processed data and experiment metadata is stored at ArrayExpress, while the raw sequence reads are held at the ENA. Complies with MIAME and MINSEQE standards. |
| Expression Atlas | EBI | Microarray RNA-Seq | Tissue-specific gene expression database for animals and plants. Displays secondary analyses and visualisation, such as functional enrichment of Gene Ontology terms, InterPro domains, or pathways. Links to protein abundance data where available. |
| Genevestigator | Privately curated | Microarray RNA-Seq | Contains manual curations of public transcriptome datasets, focusing on medical and plant biology data. Individual experiments are normalised across the full database to allow comparison of gene expression across diverse experiments. Full functionality requires licence purchase, with free access to a limited functionality. |
| RefEx | DDBJ | All | Human, mouse, and rat transcriptomes from 40 different organs. Gene expression visualised as heatmaps projected onto 3D representations of anatomical structures. |
| NONCODE | noncode.org | RNA-Seq | Non-coding RNAs (ncRNAs) excluding tRNA and rRNA. |
Legend: NCBI – National Center for Biotechnology Information;
EBI – European Bioinformatics Institute; DDBJ – DNA Data Bank of Japan;
ENA – European Nucleotide Archive; MIAME – Minimum Information About a
Microarray Experiment; MINSEQE – Minimum Information about a
high-throughput nucleotide SEQuencing Experiment.