Imagine a map of every single star in an entire galaxy. A map so detailed that it lays out what each star looks like, what they’re made of, and how each star is connected to another through the grand physical laws of the cosmos.
While we don’t yet have such an astronomical map of the heavens, thanks to a momentous study published last week in Neuron, there is now one for the brain.
If every neuron were a galaxy, then synapses—small structures dotted along the serpentine extensions of neurons—are its stars. In a technical tour-de-force, a team from the University of Edinburgh in the UK constructed the first detailed map of every single synapse in the mouse brain.
Using genetically modified mice, the team literally made each synapse light up under fluorescent light throughout the brain like the starry night. And similar to the way stars differ, the team found that synapses vastly varied, but in striking patterns that may support memory and thinking.
“There are more synapses in a human brain than there are stars in the galaxy. The brain is the most complex object we know of and understanding its connections at this level is a major step forward in unravelling its mysteries,” said lead author Dr. Seth Grant at the Center for Clinical Brain Sciences.
The detailed maps revealed a fundamental law of brain activity. With the help of machine learning, the team categorized roughly one billion synapses across the brain into 37 sub-types. Here’s the kicker: when sets of neurons receive electrical information, such as trying to decide between different solutions for a problem, unique sub-types of synapses spread out among different neurons unanimously spark with activity.
In other words: synapses come in types. And each type may control a thought, a decision, or a memory.
The neuroscience Twittersphere blew up.
“Whoa,” commented Dr. Ben Saunders simply at the University of Minnesota.
It’s an “amazing paper cataloguing the diversity and distribution of synapse sub-types across the entire mouse brain,” wrote neurogeneticist Dr. Kevin Mitchell. It “highlights [the] fact that synapses are the key computational elements in the nervous system.”
The Connectome Connection
The team’s interest in constructing the “synaptome”—the first entire catalog of synapses in the mouse brain—stemmed from a much larger project: the connectome.In a nutshell, the connectome is all the neuronal connections within you. Evangelized by Dr. Sebastian Seung in a TED Talk, the connectome is the biological basis of who you are—your memories, personality, and how you reason and think. Capture the connectome, and one day scientists may be able to reconstruct you—something known as whole brain emulation.
Yet the connectome only describes how neurons functionally talk to each other. Where in the brain is it physically encoded?
Enter synapses. Neuroscientists have long known that synapses transmit information between neurons using chemicals and electricity. There’s also been hints that synapses are widely diverse in terms of what proteins they contain, but traditionally this diversity’s been mostly ignored. Until recently, most scientists believed that actual computations occur at the neuronal body—the bulbous part of a neuron from which branches reach out.
So far there’s never been a way to look at the morphology and function of synapses across the entire brain, the authors explained. Rather, we’ve been focused on mapping these crucial connection points in small areas.
“Synaptome mapping could be used to ask if the spatial distribution of synapses [that differ] is related to connectome architecture,” the team reasoned.
And if so, future brain emulators may finally have something solid to grasp onto.
SYNMAP
To construct the mouse synaptome, the authors developed a pipeline that they dubbed SYNMAP. They started with genetically modified mice, which have their synapses glow different colors. Each synapse is jam-packed with different proteins, with—stay with me—PSD-95 and SAP102 being two of the most prominent members. The authors added glowing proteins to these, which essentially acted as torches to light up each synapse in the brain.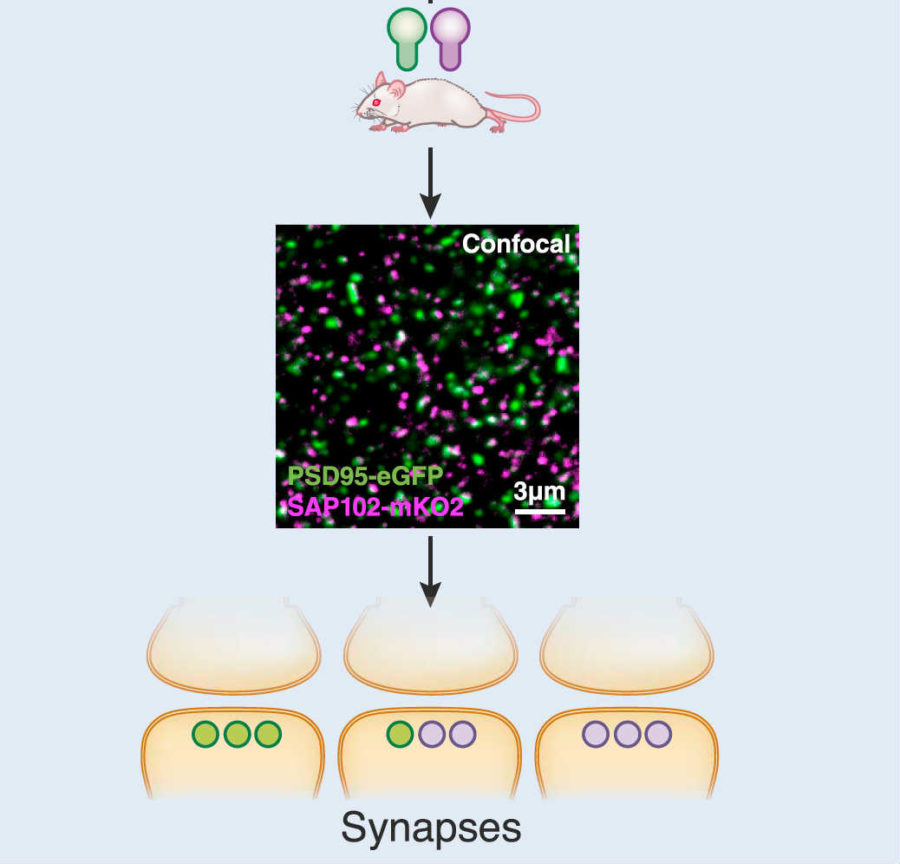
An image of synapses looks like a densely-packed star map to an untrained eye. Categorizing each synapse is beyond the ability (and time commitment) of any human researcher, so the team took advantage of new machine learning classification techniques, and developed an algorithm that could parse these data—more than 10 terabytes—automatically, without human supervision.
A Physical Connectome
Right off the bat, the team was struck by the “exquisite patterns” the glowing synapses formed. One tagged protein—PSD-95—seemed to hang out on the more exterior portions of the brain where higher cognitive functions occur. Although there is overlap, the other glowing protein preferred more interior regions of the brain.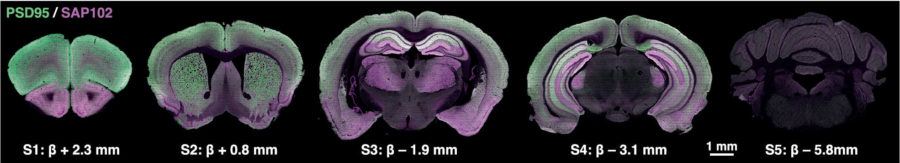
Using a machine learning algorithm developed in-house, the team categorized the synapses into 37 subtypes. Remarkably, regions of the brain related to higher reasoning and thinking abilities also contained the most diverse synapse population, whereas “reptile brain regions” such as the brain stem were more uniform in synapse sub-type.
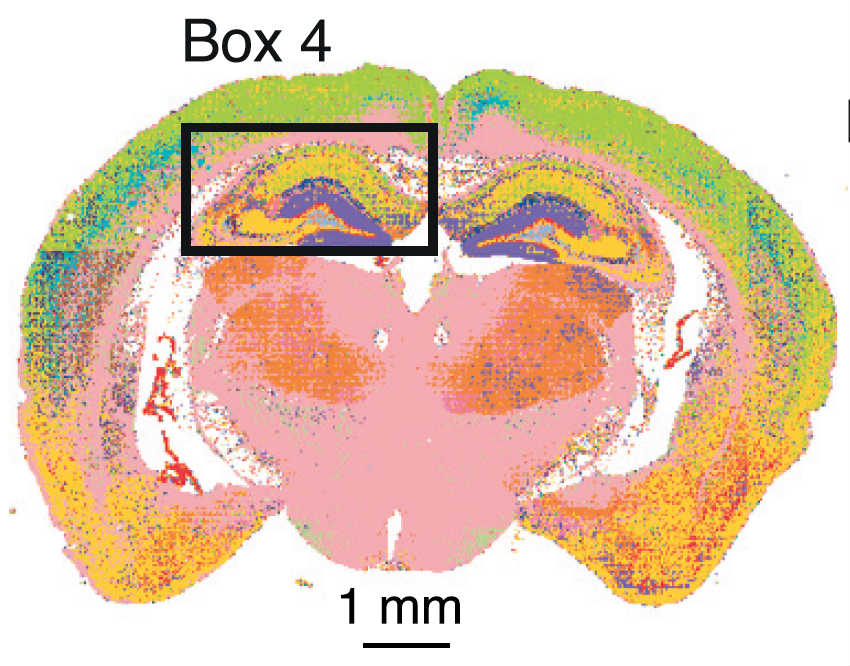
Why?
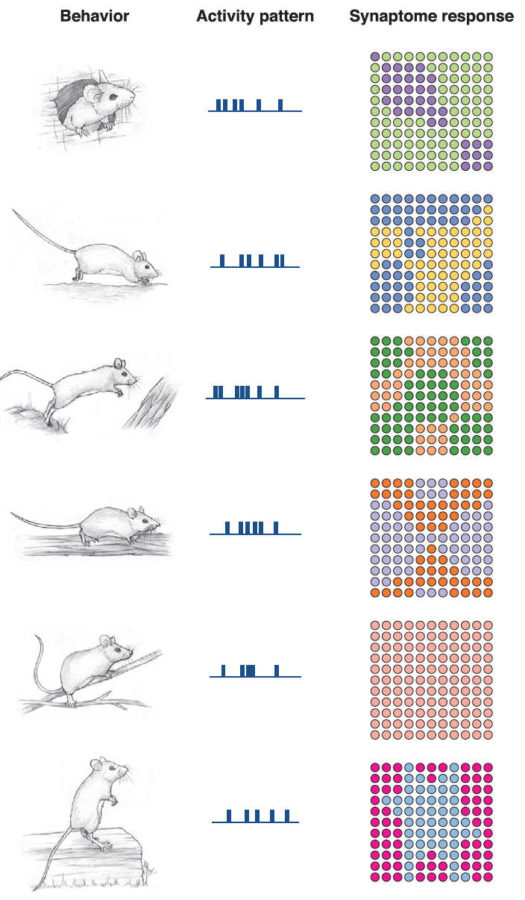
To see whether synapse diversity helps with information processing, the team used computer simulations to see how synapses would respond to common electrical patterns within the hippocampus—the seahorse-shaped region crucial for learning and memory. The hippocampus was one of the regions that showed remarkable diversity in synapse subtypes, with each spread out in striking patterns throughout the brain structure.Remarkably, each type of electrical information processing translated to a unique synaptome map—change the input, change the synaptome.
It suggests that the brain can process multiple electrical information using the same brain region, because different synaptomes are recruited.
The team found similar results when they used electrical patterns recorded from mice trying to choose between three options for a reward. Different synaptomes lit up when the choice was correct versus wrong. Like a map into internal thoughts, synaptomes drew a vivid picture of what the mouse was thinking when it made its choice.
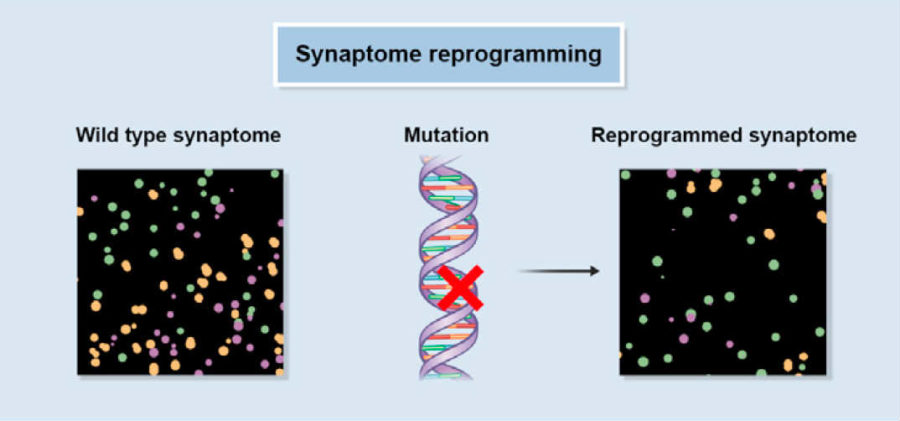
Synaptome Reprogramming
Like computer code, a synaptome seems to underlie a computational output—a decision or thought. So what if the code is screwed up?Psychiatric diseases often have genetic causes that impact proteins in the synapse. Using mice that show symptoms similar to schizophrenia or autism, the team mapped their synaptome—and found dramatic changes in how the brain’s various synapse sub-types are structured and connected.
For example, in response to certain normal brain electrical patterns, some synaptome maps only weakly emerged, whereas others became abnormally strong in the mutant mice.
So are you your synaptome?
Perhaps. The essence of you—memories, thought patterns—seems to be etched into how diverse synapses activate in response to input. Like a fingerprint for memories and decisions, synaptomes can then be “read” to decipher that thought.But as the authors acknowledge, the study’s only the beginning. Along with the paper, the team launched a Synaptome Explorer tool to help neuroscientists further parse the intricate connections between synapses and you.
“This map opens a wealth of new avenues of research that should transform our understanding of behavior and brain disease,” said Grant.
Images Credit: Derivatives of Fei Zhu et al. / University of Edinburg / CC BY 4.0
